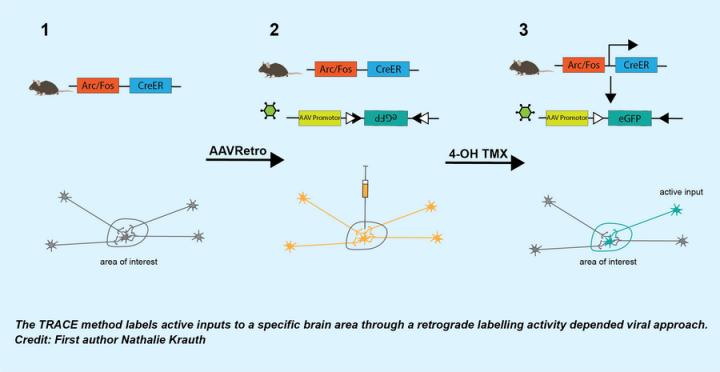
Credit: Nathalie Krauth
A major interest in brain research is understanding behavioural experiences at the synaptic and circuit level. However, gaining insight into the neural circuits that underlie a specific behavioural experience is a challenge. The perplexity lies in defining which of the large number of neuronal inputs a specific region of the brain receives, expresses a defined behaviour, and yet there are no direct methods available to overcome this challenge. Today, a common mapping approach in neuroscience is to use retrograde tracing viruses and identify all neuronal inputs in a population of neurons in a specific region of the brain. But neuroscientists are still left to rely on their experience, knowledge and previous findings to exactify specific inputs that underlie a defined behaviour.
A collaborative study led by circuitry experts at the department of Molecular Biology and Genetics, department of Biomedicine and centre of Proteins in Memory unravel a new approach or so to say, an unbiased and accurate strategy, that selectively tags arriving inputs that are activated by a defined stimulus and projected to a particular brain region. The researchers’ new approach (TRACE) uses an approach where a retrograde virus is combined with a Cre-recombinase that is under the control of an activity dependent promoter to express a fluorescent marker in input neurons.
Using mouse models, the Nabavi and Capogna group have demonstrated the input specificity of their novel approach on four different brain circuits receiving defined sensory inputs. Researchers performed behavioural testing using optogenetic stimulation, odor driven innate fear detection and shock-induced circuit experiments. They also performed imaging of brain slices to investigate specific neural activity.
Their findings show that TRACE can identify active sensory neural inputs that underlie a defined behaviour. This new labelling system is an additional tool in the toolbox to gain access to neurons in the brain that are activated by a particular stimulation or experience.
###
The publication can be read in full at the Frontiers in Cellular Neuroscience website: https:/
Learn more about the Capogna group and their research within amygdala-hippocampal-cortical circuits in health and disease: https:/
Visit the Nabavi group webpage to learn more their research within memory formation and consolidation at the synaptic and circuit level: https:/
Media Contact
Sadegh Nabavi
[email protected]
Original Source
https:/
Related Journal Article
http://dx.