New theory uses physics to predict the growth stages of cancerous tumors
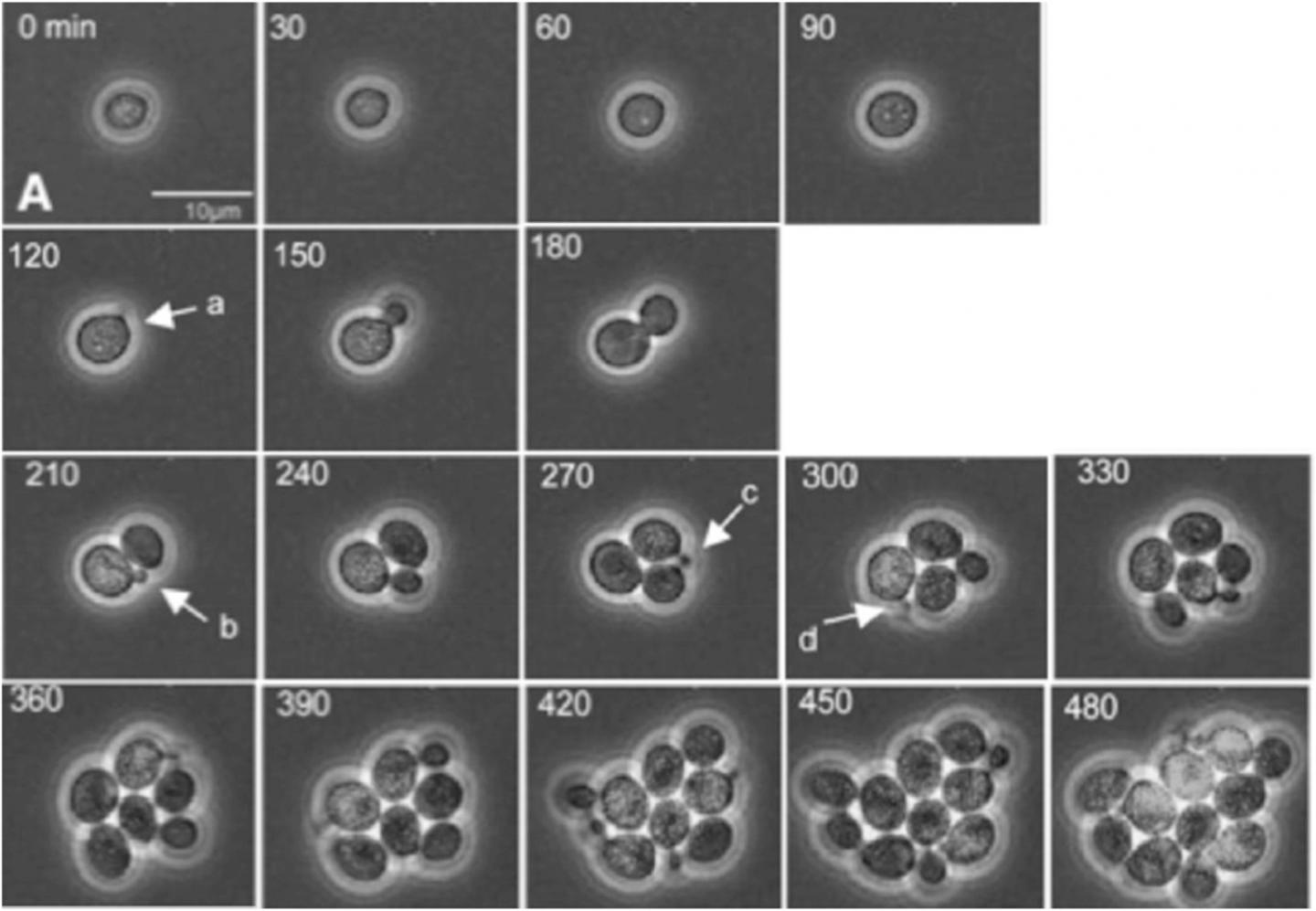
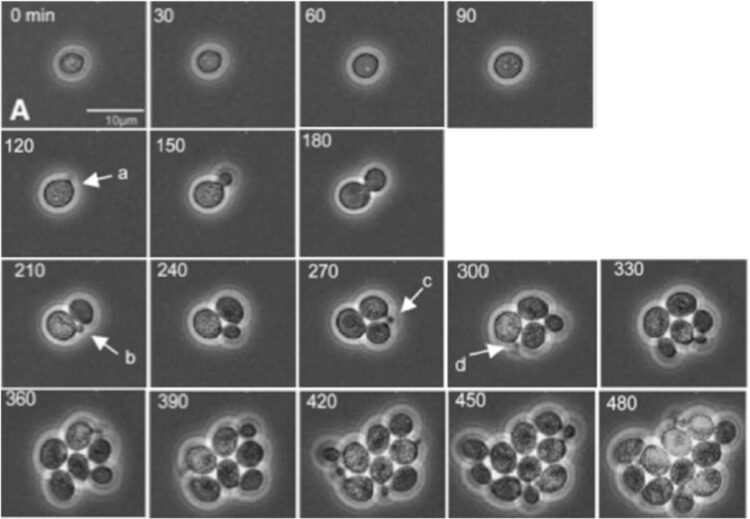
Credit: Bean, et al. Mol. Cell, 2006. DOI: 10.1016/j. molcel.2005.10.035.
DURHAM, N.C. — Researchers at Duke University have developed a predictive theory for tumor growth that approaches the subject from a new point of view. Rather than focusing on the biological mechanisms of cellular growth, the researchers instead use thermodynamics and the physical space the tumor is expanding into to predict its evolution from a single cell to a complex cancerous mass.
The results appeared Jan. 15 in the journal Biosystems.
“When scientists think about cancer, the first thing that comes to mind is biology, and they tend to overlook the physical reality of its growth pattern,” said Ehsan Samei, professor of radiology at the Duke University School of Medicine. “There are also issues of how a tumor interacts with its surroundings and provides nutrients to its cells, which is an aspect of the story that so far hasn’t been focused on very much.”
“Our theory uses fundamental concepts of how natural systems grow and reorganize themselves to gain greater access to flowing systems,” said Adrian Bejan, the J.A. Jones Distinguished Professor of Mechanical Engineering at Duke. “It not only explains how this growth should happen, but also why it must happen.”
The new work is based on the constructal law, which Bejan penned in 1996, that states that for a system to survive, it must evolve to increase its access to flow. For example, the human vascular system has evolved to provide blood flow through a network of a few large arteries and many small capillaries. River systems, tree branches and modern highway and road networks all reflect the same forces at work.
In the paper, Bejan, Samei and doctoral student Thomas Sauer demonstrate how a tumor’s growth and internal reorganization as it grows are directly tied to its need to create greater access to flowing nutrients as well as conduits for removing refuse. They use these insights to predict the growth of cell clusters as a function of structure, and also to predict the critical cluster sizes that mark the transitions from one distinct configuration to the next.
To validate their theory, the researchers compared their predictions with the measurements of several independent studies of cancerous and non-cancerous tumor growth patterns. The results show that their work provides a unifying perspective on the growth of cell clusters on the smallest scales as well as the large-scale dynamics of proliferating cells as described in phenomenological models.
“As the tumor grows, the flow systems get large enough to create visible currents through the vascularization of the tumor,” said Bejan. “Our theory reveals the physics behind these sorts of dramatic transitions and predict when they should happen.”
For the past 15 years, Samei has been working on computational, extremely detailed models of patients to test new interventions and medical procedures through so-called virtual clinical trials. These trials emulate the patient and the procedures to enable medical evaluations that would otherwise be prohibitively costly or unethical. Until recently, the tumors represented in these virtual patients have been static. But with the new theory of tumor growth, Samei is successfully creating dynamic, growing tumor models that may open new pathways to cancer research.
Samei is currently in the process of generating tumor growth images from his new models, mixing them with images taken from real patients, and testing radiologists to see if they can tell the difference. So far, the results have been promising.
“If the tumors don’t grow, the model isn’t realistic,” said Samei. “We want our platform to be able to accurately model tumor growth so that we can answer the very real question of how often a patient needs to be imaged to determine whether or not their cancer is growing or shrinking.”
###
CITATION: “Cell and Extracellular Matrix Growth Theory and its Implications for Tumorigenesis,” T.J. Sauer, E. Samei, A. Bejan. Biosystems, Jan. 15, 2021. DOI: 10.1016/j.biosystems.2020.104331
Media Contact
Ken Kingery
[email protected]
Original Source
https:/
Related Journal Article
http://dx.