Credit: Chung Lab/MIT
When there’s a vexing problem to be solved, people sometimes offer metaphorical advice such as “stretching the mind” or engaging in “flexible” thinking, but in confronting a problem facing many biomedical research labs, a team of MIT researchers has engineered a solution that is much more literal. To make imaging cells and molecules in brain and other large tissues easier while also making samples tough enough for years of handling in the lab, they have come up with a chemical process that makes tissue stretchable, compressible and pretty much indestructible.
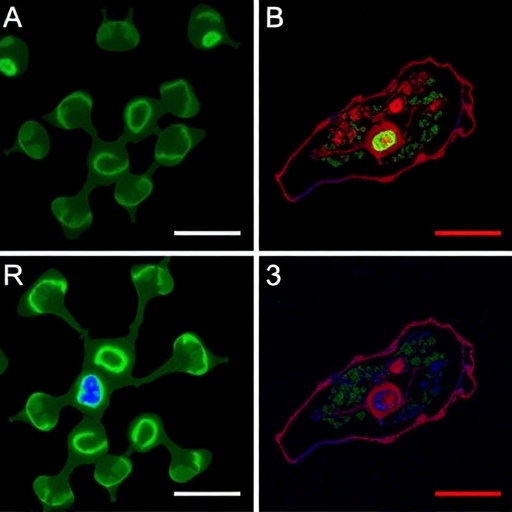
“ELAST” technology, described in a new paper in Nature Methods, provides scientists a very fast way to fluorescently label cells, proteins, genetic material and other molecules within brains, kidneys, lungs, hearts and other organs. That’s because when such tissues can be stretched out or squished down thin, labeling probes can infuse them far more rapidly. Several demonstrations in the paper show that even after repeated expansions or compressions to speed up labeling, tissues snap back to their original form unaltered except for the new labels.
The lab of Kwanghun Chung, an associate professor of chemical engineering and a member of MIT’s Institute for Medical Engineering and Science, and Picower Institute for Learning and Memory, developed ELAST amid work on a five-year project, funded by the National Institutes of Health, to make the most comprehensive map yet of the entire human brain. That requires being able to label and scan every fine cellular and molecular detail in the thickest slabs possible to preserve 3D structure. It also means the lab must be able to keep samples perfectly intact for years, even as they must accomplish numerous individual rounds of labeling quickly and efficiently. Each round of labeling – maybe a particular kind of neuron one day, or a key protein the next – will tell them something new about how the brain is structured and how it works.
“When people donate their brain, it is like they are donating a library,” said Chung. “Each one contains a library worth of information. You cannot access all the books in the library at the same time. We have to repeatedly be able to access the library without damaging it. Each of these brains is an extremely precious resource.”
Former lab postdoc Taeyun Ku, now an assistant professor at the Korea Advanced Institute of Science and Technology, is the study’s lead author. He said the particular difficulty of working with human tissues, which of course are much larger than those of lab animals like mice, inspired him to take this new engineering approach. Late one night in the lab around Christmas 2017, he said, he was mulling over how to transform tissue for quicker labeling and began to tinker with repeated compression of an elastic gel.
“We changed our way of thinking: biological tissue doesn’t need to be very biological,” Ku said. “If our goal is not to image living events but to image appearances, we can change the material type of the tissue while maintaining the appearances. Our work shows how higher-level engineering of the brain enables us to better look into what inside the brain.”
Entangled links
The team’s efforts to engineer ELAST came down to finding the right formulation of a gel-like chemical called polyacrylamide. In the past Chung has used the substance in a different formulation with crosslinking chemicals to make tissues strong but fairly brittle, said study co-author Webster Guan, a chemical engineering graduate student. When that formulation infused the tissues, cells and molecules would become directly attached to a grid-like mesh.
In the new formulation, the team used a high concentration of acrylamide with much less crosslinker and initiator. The result was an entanglement of long polymer chains with links that are able to slip around, giving the gel a structural integrity but with much more flexibility. Moreover, rather than attaching to the chains, Guan said, the cells and molecules of the tissue just become entangled within it, adding further to the ability of the acrylamide-infused tissues to withstand stretching or squashing without anything becoming torn or permanently displaced in the process.
In the study the team reports stretching human or mouse brain tissues to twice their width and length simultaneously or compressing their thickness by 10 times with virtually no distortion after returning to their regular size.
“These results demonstrate that ELAST enables fully reversible tissue shape transformation while preserving structural and molecular information in the tissue,” they wrote.
Fully integrating the polyacrylamide into a large amount of tissue to achieve the elasticity can take as long as 21 days, they report, but from then on, any individual labeling step, such as labeling a particular kind of cell to determine its abundance, or a specific protein to see where it is expressed, can proceed far more quickly than with prior methods.
In one case, by repeatedly compressing a 5-milimeter thick cross section of a human brain, the team needed only 24 hours to label it all the way through. For comparison, back in 2013 when Chung and colleagues debuted “CLARITY,” a method of making brain tissue transparent and fixing it with an acrylamide gel, they needed 24 hours to label a slice only a tenth as thick. Because labeling time is estimated by squaring the depth that probes must penetrate, calculations suggest labeling with ELAST proceeds 100 times faster than with CLARITY.
Though Chung’s lab mostly focuses on brains, the applicability to other organs can aid in other cell mapping efforts, Chung said. He added that even if labeling tissue isn’t a goal at all, having an easy new way to make a durable, elastic gel could have other applications, for instance in creating soft robotics. Resources for learning more about ELAST are available at Chung’s website.
###
In addition to Ku, Guan and Chung, the paper’s other authors are Nicholas Evans, Chang Ho Sohn, Alexandre Albanese, Joon-Goon Kim, and Matthew Frosch, professor at Massachusetts General Hospital and Harvard Medical School.
Funding for the work came from sources including the JPB Foundation, the NIH, the NCSOFT Cultural Foundation, Searle Scholars Program, Packard award in Science and Engineering, NARSAD Young Investigator Award, and McKnight Foundation Technology Award.
Media Contact
David Orenstein
[email protected]
Related Journal Article
http://dx.