The novel biosensor developed by scientists at Rice University in collaboration with colleagues in Thailand and Ireland may make the detection of pathogens much faster and easier for food-manufacturing plants.
A study on the discovery appears online this month in the American Chemical Society journal Analytical Chemistry.
The process appears to easily outperform tests that are now standard in the food industry. The standard tests are slow because it can take days to culture colonies of salmonella bacteria as proof, or laborious because of the need to prepare samples for DNA-based testing.
The Rice process delivers results within minutes from a platform that can be cleaned and reused. The technology can be easily customized to detect any type of bacteria and to detect different strains of the same bacterium, according to the researchers.
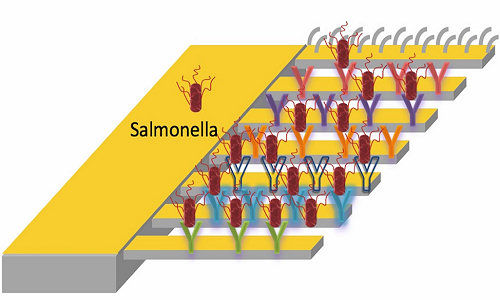
The “diving boards” are a set of microcantilevers, each of which can be decorated with different peptides that have unique binding affinities to strains of the salmonella bacteria. When a peptide catches a bacterium, the cantilever bends ever so slightly, due to a mismatch in surface stress on the top and bottom. A fine laser trained on the mechanism catches that motion and triggers the alarm.
The system is sensitive enough to warn of the presence of a single pathogen, according to the researchers, who wrote that very low pathogen concentrations cause foodborne disease.
The idea springs from research into the use of microcantilevers by Rice biomolecular engineer Sibani Lisa Biswal and lead author Jinghui Wang, a graduate student in her lab. Biswal was prompted to have a look at novel peptides by her graduate school friend, Nitsara Karoonuthaisiri, head of the microarray laboratory at the National Center for Genetic Engineering and Biotechnology in Thailand. Karoonuthaisiri is also a visiting scientist at the Institute for Global Food Security at the Queen’s University, Belfast.
“She’s been working in this area of pathogenic bacteria and asked if we have thought about trying to use our microcantilevers for detection,” Biswal said. “Specifically, she wanted to know if we could try these novel peptides.”
Karoonuthaisiri and her team had isolated bacteriophage viruses associated with salmonella through biopanning and phage display, a technique to study interactions among proteins, peptides and pathogens. She then derived peptides from the phages that would serve as targets for specific bacteria.
“She said, ‘We spend a lot of time trying to characterize which of these peptides work the best. It looks like you have a platform that can do and quantitate that.’ So that’s where we came in,” Biswal said.
The Rice lab compared the peptides’ performance with commercial antibodies now used for salmonella detection and found the peptides were not only more sensitive but could be used in a multiplexed cantilever array to detect many different kinds of salmonella at once.
“The peptides are very robust,” Biswal said. “That’s why a lot of people like them over antibodies. The peptides can handle harsher conditions and are much more stable. Antibodies are large proteins and break down more readily.
“We’re very excited to see where this will lead,” she said.
Story Source:
The above story is based on materials provided by Rice University.
Publication: Rapid Detection of Pathogenic Bacteria and Screening of Phage-Derived Peptides Using Microcantilevers. Jinghui Wang, M. Josephine Morton, Christopher T. Elliott, Nitsara Karoonuthaisiri, Laura Segatori, and Sibani Lisa Biswal. Analytical Chemistry (Article ASAP): http://pubs.acs.org/doi/abs/10.1021/ac403437x