Terminological confusion between species
Credit: UNIVERSITY OF BARCELONA
Oxytocin and arginine vasopressin are two hormones in the endocrine system that can act as neurotransmitters and regulate -in vertebrates and invertebrates- a wide range of biological functions, such as bonding formation, breastfeeding, birth or arterial pressure. Biochemists in the pregenomic era, named these genes differently in different species, due to small protein coding differences.
A new study carried out by the University of Barcelona (UB) and the Rockefeller University, published in the journal Nature, has analysed and compared the genome of 35 species representing all vertebrate lineages and reached the conclusion that both hormones stem from a common ancestral gene.
These high-quality genomes were obtained thanks to the use of innovative genome sequencing technologies in the context of the Vertebrate Genomes Project. Based on these findings, the researchers propose a new universal nomenclature, based on their identified evolutionary history for these genes. According to the authors of the study, the proposal allows for a new nomenclature that is universal across vertebrates, both for the hormone-genes and their receptors, therefore easing the species comparative research.
The paper is part of the doctoral thesis by Constantina Theofanopoulou, carried out at the UB under the co-supervision of Cedric Boeckx, ICREA researcher at the Institute of Complex Systems of the UB (UBICS) and Erich D. Jarvis, professor at the Rockefeller University.
Terminological confusion between species
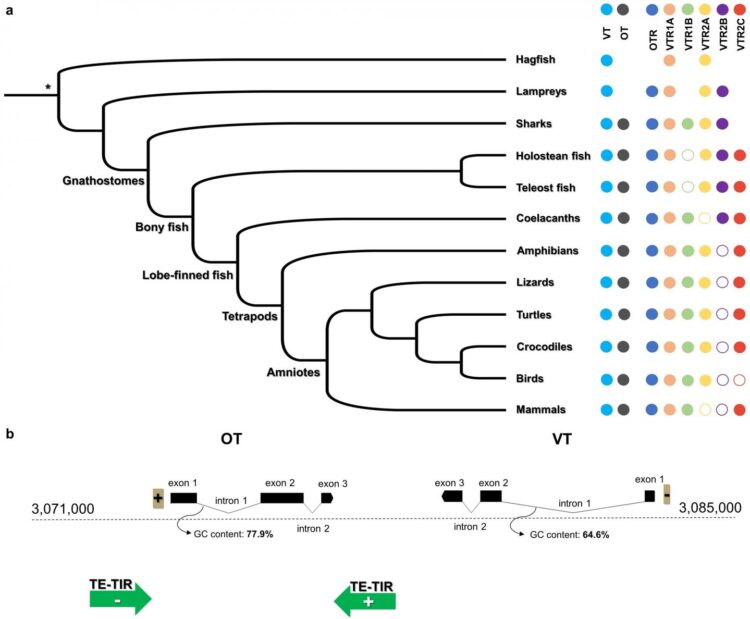
The study results from problems the first author of the study, Constantina Theofanopoulou, became aware of in the literature of these genes, when analysing the role of oxytocin in the vocal learning ability in songbirds. “When analysing the scientific literature to find previous work on this gene, we observed that when talking about birds, scientists used the term ‘mesotocin’, instead of ‘oxytocin’. Different names, like ‘isotocin’ or ‘neurophysin’ were used in other vertebrate species, such as turtles, frogs or fish. An equally confusing nomenclature was the one used for their respective receptors, making it very difficult to understand which gene is which in different species and lineages”, says Constantina Theofanopoulou. In this context, the researchers aimed to find out which genes were truly orthologous to each other in different species. To do so, they used the genome assemblies generated by the Vertebrates Genome Project, an international collaboration led by Erich Jarvis, from the Rockefeller University. In the first phase, Theofanopoulou and the research team compared the genomes and specifically the order of genes around their genes of interest. They studied up to sixty genes around the genes of interest, as well as all the genes of the chromosomes where these genes are located. They sought to find out whether the order of genes -called synteny- was conserved across species, something that would shed light to the evolutionary history of these genes.
The new high-quality assemblies were more than helpful since, as Theofanopoulou says, “Most genomes so far have had low quality, meaning millions of mistakes, parts of genes being absent or incorrectly assembled. These errors can have direct consequences on the scientific findings”.
Genes with a common ancestor
The results, together with other phylogenetic analyses carried out during the study, show that genes that the researchers of this study now call oxytocin and vasotocin -that code for the homonymous hormones- are paralogous, that is, they come from a local duplication of a common ancestral gene. “Our findings suggest that the oxytocin gene is a duplication of the vasotocin gene -historically called vasopressin- that took place after the divergence of jawed vertebrates from jawless vertebrates. This simultaneously means that, if we had understood the evolution of these genes from the very beginning, we would most likely not have used different names to refer to them, as we see in other gene families, where we use the same root name for all its gene-members (e.g., FOXP) and different numbers to differentiate them (e.g., FOXP1, FOXP2 etc.)”, highlights Constantina Theofanopoulou.
Given the results, the researchers propose a universal nomenclature in which oxytocin and vasotocin are used for these genes in all jawed vertebrates, and vasotocin for the only gene present all jawless vertebrates and closely related invertebrates. In this proposal, the common origin of both genes would be represented through the shared suffix -tocin, while the parology would be displayed through the different prefixes (oxy- and ¬vaso-). Following this new proposed nomenclature, what is currently called oxytocin, mesotocin, isotocin, glumitocin, valitocin, aspargtocin and neurophysin in different lineages, would be now called oxytocin universally. As for what is currently named vasotocin, vasopressin, neurophysin 2 and phenypresin in different species, would now be called vasotocin for all species.
This proposal also includes a universal nomenclature for the receptors of both genes, being oxytocin and vasotocin receptors (OTR and VTRs). Therefore, what has traditionally been called oxytocin receptor (OXTR) in mammals, vasotocin 3 receptor (VT3) and mesotocin receptor (MTR) in birds and frogs, and isotocin receptor (ITR) in fish, would be now called oxytocin receptor (OTR) in all cases.
Evolutionary history of oxytocin and vasotocin
The study also identified six main receptors of oxytocin and vasotocin in jawed vertebrates, with two of them being present already in the hagfish, two additional in lampreys, which have four receptors in total, and two more in jawed, which display six receptors in total. This pattern suggests that these receptors have first evolved through one round of whole genome duplication that gave rise to the four receptors we find in jawless vertebrates, like the lampreys, and that afterwards either smaller-scale segmental duplications or another whole genome duplication gave rise to the six receptors in jawed vertebrates.
This is the second time researchers find evidence for a scenario with one round of whole genome duplication, instead of two. The hypothesis that vertebrate genomes have evolved through two rounds of whole genome duplication has not been challenged in almost fifty years. “Our data show more support for the one round of whole genome duplication-idea. If the two rounds of whole genome duplication were the case, we would expect to find eight in total and not six receptors in jawed vertebrates. Of course, gene loss is very common, so that’s why we can’t exclude a two round-scenario, but we did not find any genomic sign of gene loss in any of the jawed vertebrate genomes we studied”, notes the researcher.
The next step would be to get this new nomenclature, which is based on the evolutionary history of genes, adopted. “This is the first time that such a universal change affecting so many genes and species is being presented. Regardless of whether this proposal will be adopted or not, researchers will now have a guide on the genetic orthologies among species, which will make their research much easier”, concludes Constantina Theofanopoulou.
###
Media Contact
Rosa Martínez
[email protected]
Original Source
https:/
Related Journal Article
http://dx.