Review of the life histories of RNA viruses, their genetics, and the compounds that could disrupt them offers hope for curtailment of future pandemics
Credit: Illustration: Jennifer Matthews/Scripps Oceanography
Researchers at Scripps Institution of Oceanography and Skaggs School of Pharmacy and Pharmaceutical Sciences at the University of California San Diego have broken down the genomic and life history traits of three classes of viruses that have caused endemic and global pandemics in the past and identify natural products – compounds produced in nature – with the potential to disrupt their spread.
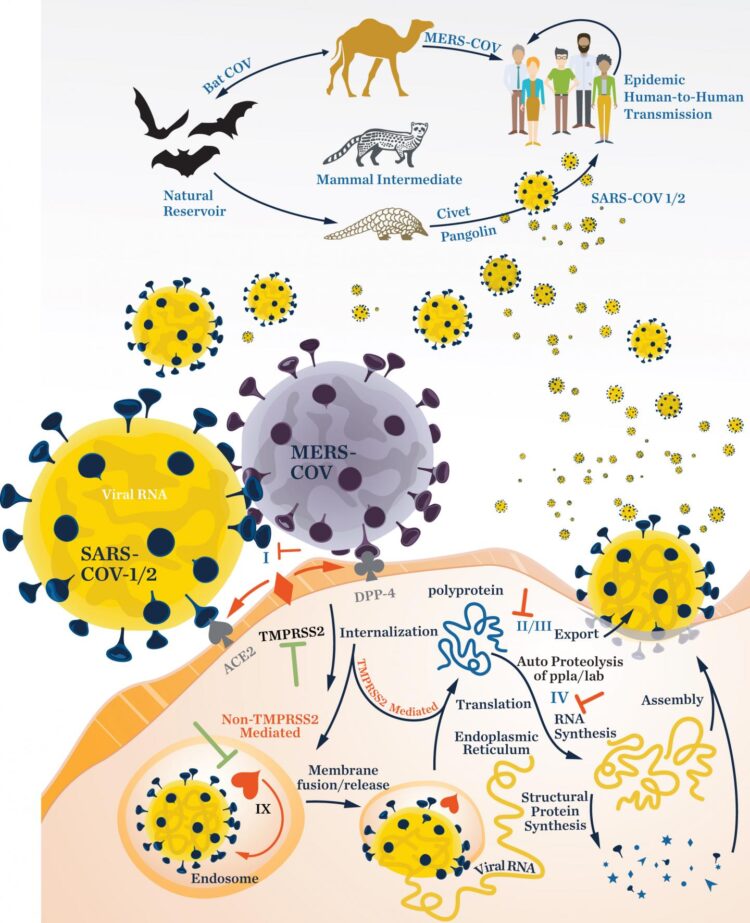
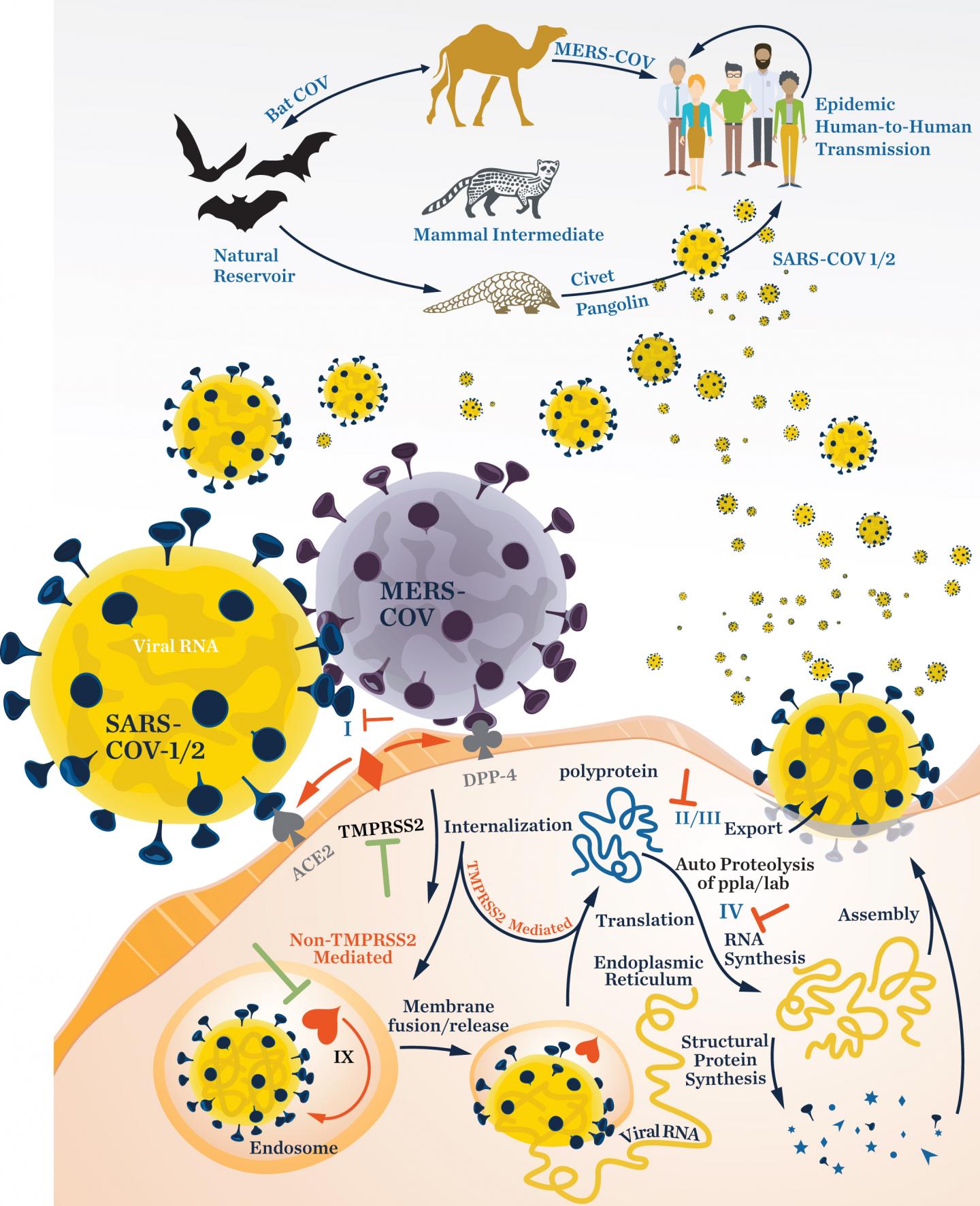
In a review appearing in the Journal of Natural Products, marine chemists Mitchell Christy, Yoshinori Uekusa, and William Gerwick, and immunologist Lena Gerwick describe the basic biology of three families of RNA viruses and how they infect human cells. These viruses use RNA instead of DNA to store their genetic information, a trait that helps them to evolve quickly. The team then describes the natural products that have been shown to have capabilities to inhibit them, highlighting possible treatment strategies.
“We wanted to evaluate the viruses that are responsible for these deadly outbreaks and identify their weaknesses,” said Christy, the first author. “We consider their similarities and reveal potential strategies to target their replication and spread. We find that natural products are a valuable source of inhibitors that can be used as a basis for new drug development campaigns targeting these viruses.”
The research team is from Scripps Oceanography’s Center for Marine Biotechnology and Biomedicine (CMBB), which collects and analyzes chemical compounds found in marine environments for potential efficacy as antibiotics, anticancer therapies, and other products with medical benefit. A drug known as Marizomib entered the final stages of clinical trials as a potential treatment for brain cancers earlier in 2020. The drug came from a genus of marine bacteria that CMBB researchers had originally collected in seafloor sediments in 1990.
The researchers, funded by the National Institutes of Health and the UC San Diego Chancellor’s Office, present an overview of the structure of viruses in the families Coronaviridae, Flaviviridae, and Filoviridae. Within these families are viruses that have led to COVID-19, dengue fever, West Nile encephalitis, Zika, Ebola, and Marburg disease outbreaks. The team then identifies compounds produced by marine and terrestrial organisms that have some demonstrated level of activity against these viruses. Those compounds are thought to have molecular architectures that make them potential candidates to serve as viral inhibitors, preventing viruses from penetrating healthy human cells or from replicating. The goal of the review, the researchers said, was to improve the process of drug development as new pandemics emerge, so that containing disease spread can accelerate in the face of new threats.
“It is simply common sense that we should put into place the infrastructure necessary to more rapidly develop treatments when future pandemics occur,” the review concludes. “One such recommendation is to create and maintain international compound libraries with substances that possess antiviral, antibacterial, or antiparasitic activity.”
To achieve that goal, the researchers realize that international agreements would need to be reached to address intellectual property issues, the rights and responsibilities of researchers, and other complex issues.
And while there has been remarkable progress in the development of vaccines for SARS-CoV-2 infection, effective antiviral drugs are also critically needed for managing COVID-19 infection in unvaccinated individuals or in cases where the efficacy of a vaccine decreases over time, the researchers said. While several candidate antiviral molecules have been investigated for use in the clinic, such as remdesivir, lopinavir-ritonavir, hydroxychloroquine, and type I interferon therapy, all have shown limited or no efficacy in large scale trials. Effective antiviral drugs are still much in need of discovery and development.
###
Media Contact
Robert Monroe
[email protected]
Original Source
https:/
Related Journal Article
http://dx.