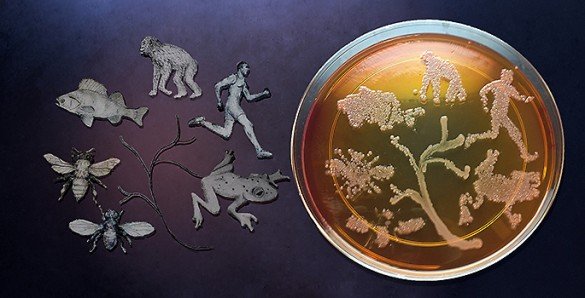
You are not just yourself. You are also the thousands of microbes that you carry. In fact, they represent an invisible majority that may be more you than you realize.
These microscopic fellow travelers are collectively called the microbiome. Realization that every species of plant and animal is accompanied by a distinctive microbiome is old news. But evidence of the impact that these microbes have on their hosts continues to grow rapidly in areas ranging from brain development to digestion to defense against infection to producing bodily odors.
Now, contrary to current scientific understanding, it also appears that our microbial companions play an important role in evolution. A new study, published online on July 18 by the journal Science, has provided direct evidence that these microbes can contribute to the origin of new species by reducing the viability of hybrids produced between males and females of different species.
This study provides the strongest evidence to date for the controversial hologenomic theory of evolution, which proposes that the object of Darwin’s natural selection is not just the individual organism as he proposed, but the organism plus its associated microbial community. (The hologenome encompasses the genome of the host and the genomes of its microscopic symbiotes.)
“It was a high-risk proposition. The expectation in the field was that the origin of species is principally driven by genetic changes in the nucleus. Our study demonstrates that both the nuclear genome and the microbiome must be considered in a unified framework of speciation,” said Associate Professor of Biological Sciences Seth Bordenstein who performed the study with graduate student Robert Brucker.
They conducted their research using three species of the jewel wasp Nasonia. These tiny, match-head sized wasps parasitize blowflies and other pest flies, which make them useful for biological control.
“The wasps have a microbiome of 96 different groups of microorganisms,” said Brucker. Two of the species they used (N. giraulti and N. longicornis) only diverged about 400,000 years ago so they are closely related genetically. This closeness is also reflected in their microbiomes, which are quite similar. The third species (N. vitripennis), on the other hand, diverged about a million years ago so there are greater differences in both its genome and microbiome, he explained.
The mortality of hybrid offspring from the two closely related species was relatively low, about 8 percent, while the mortality rate of hybrid offspring between either of them and N. vitripennis was quite high, better than 90 percent, the researchers established.
“The microbiomes of viable hybrids looked extremely similar to those of their parents, but the microbiomes of those that did not survive looked chaotic and totally different,” Brucker reported.
The researchers showed that the incompatibilities that were killing the hybrids had a microbial basis by raising the wasps in a microbe-free environment. They were surprised to find that the germ-free hybrids survived just as well as purebred larvae. But when they gave the germ-free hybrids gut microbes from regular hybrids, their survival rate plummeted.
“Our results move the controversy of hologenomic evolution from an idea to an observed phenomenon,” said Bordenstein. “The question is no longer whether the hologenome exists, but how common it is?”
This research was supported by award DEB 1046149 from the National Science Foundation’s Dimensions of Biodiversity program.
Story Source:
The above story is reprinted from materials provided by Vanderbilt University, David Salisbury.