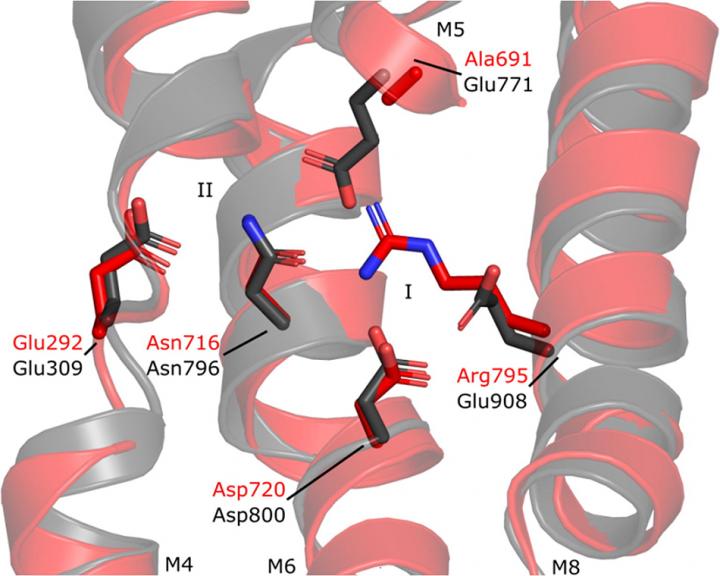
Many bacteria export intracellular calcium using active transporters homologous to the well-described mammalian Ca2+-ATPases such as plasma-membrane Ca2+-ATPase and sarco-endoplasmic reticulum Ca2+-ATPase (PMCA and SERCA, respectively). Crystal structures of Ca2+-ATPase 1 from Listeria monocytogenes (LMCA1) suggest that LMCA1 is pre-organized for dephosphorylation upon Ca2+ release, which can explain the rapid dephosphorylation observed earlier in single-molecule studies.
Also, variation in the architecture of the calcium binding sites explains why LMCA1 transports a single Ca2+ ion similar to PMCA, in contrast to two transported Ca2+ ions in SERCA. The LMCA1 structures provide insight into the evolutionary divergence and conserved features of this important class of ion transporters that also inform us on central mechanisms of mammalian Ca2+ -ATPases and how they can be regulated or affected by pathological conditions.
For the P4-ATPase study, researchers took a different perspective. The transport cycle of a P-type ATPase consist of two half-reactions. Phosphorylation where a phosphate is transferred from ATP to the transporter, and dephosphorylation, where the phosphate is again released. In contrast to ion transporters such as LMCA1, the P4-ATPases transport lipids and are known as lipid flippases. Importantly, the lipid transport is coupled to the dephosphorylation reaction of the cycle, where for ion transporting P-type ATPases it is mainly coupled to the phosphorylation reaction.
Through new structures determined by cryo-electron microscopy (cryo-EM) of a yeast lipid flippase, Drs2p/Cdc50p, it was investigated how the lipid flippases have diverged from ion transporters and have adapted the enzymatic mechanism for the “flipped” purpose. Cryo-EM was a critical technique for this study, and multiple structures of the transport cycle could be determined by locking Drs2p/Cdc50p using different inhibitors and electron microscopy data collected at the electron microscopy infrastructure facility at Aarhus University (EMBION).
The two studies have been spearheaded by PhD student Sara Basse Hansen and Postdoc Milena Timcenko – under the supervision of Professor Poul Nissen (and Sara also of Associate Professor Magnus Kjærgaard) – and are being published in Journal of Molecular Biology.
###
The results have been published in two articles in Journal of Molecular Biology:
The Crystal Structure of the Ca2+-ATPase 1 from Listeria monocytogenes reveals a Pump Primed for Dephosphorylation
Sara Basse Hansen, Mateusz Dyla, Caroline Neumann, Esben Meldgaard Hoegh Quistgaard, Jacob Lauwring Andersen, Magnus Kjaergaard & Poul Nissen
Structural basis of substrate-independent phosphorylation in a P4-ATPase lipid flippase: Milena Timcenko, Cédric Montigny, Thomas Boesen, Joseph A. Lyons, Guillaume Lenoir & Poul Nissen
Media Contact
Professor Poul Nissen
[email protected]
Original Source
https:/
Related Journal Article
http://dx.