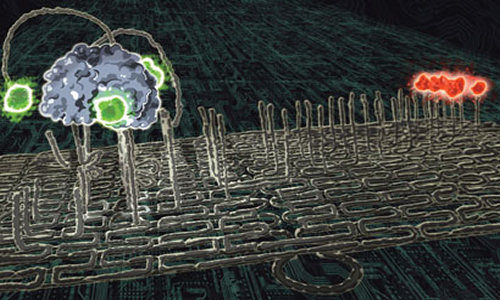
With the relentless rise of DNA nanotechnology’s popularity, Emma Davies explores the role chemistry has played in its success
As a supramolecular chemist, Hanadi Sleiman found herself strongly drawn to manmade DNA structures. ‘We think of DNA as the most programmable structure there is. I thought – if it is – let me try to incorporate it into regular supramolecular structures,’ says the professor at McGill University, Montreal, Canada. She hasn’t looked back. ‘What is really beautiful about DNA structures is the fact that you can control every single aspect of them,’ she exclaims.
Sleiman is one of an increasing number of chemists who have turned to DNA nanotechnology. Some pin their hopes on using DNA in nanoelectronics or for drug delivery, while others are excited about its potential as an analytical tool.
Single strands of DNA bind (hybridise) to each other when the bases cytosine and guanine (C-G) and adenine and thymine (A-T) couple through hydrogen bonds and a good geometric fit. The fact that a single strand of DNA will only bind to another with the correct base sequence gives researchers incredible structure control. Meanwhile, fully automated DNA synthesisers mean that DNA sequences are easy to construct.
In 2006 Paul Rothemund, a computational bioengineer at the California Institute of Technology in the US published a paper outlining a strikingly simple way to create wonderful patterned templates using DNA strands – coining the phrase ‘DNA origami’. Long, single DNA strands form a template while hundreds of shorter DNA ‘staple’ strands hold the template together and force it to fold – like origami – into a series of parallel lines within the desired shape outline. The DNA sections can be bought commercially and construction is done at room temperature. Andrew Turberfield, a physicist who specialises in DNA nanotechnology at the University of Oxford, UK, has described this technology as triggering ‘a small revolution in DNA self-assembly’.
‘DNA origami really works,’ agrees Friedrich Simmel, a biophysicist at the Technical University of Munich in Germany. ‘What is amazing and exciting for many people is that when you mix the DNA sequences in the lab the origami works almost immediately.’ In 2011 Rothemund reported a method to extend the size of DNA structures that can be created by slotting a series of origami tiles together using base pair stacking between G-C and C-G base pairs on the edge of tiles.
Super supramolecules
While McGill’s Sleiman describes Rothemund’s original origami as a ‘beautiful discovery’, she takes a different approach to DNA structure building. Her team has added a chemical slant, building 3D DNA structures with synthetic molecules as key components. ‘We use a lot of synthetic molecules that we embed in the DNA. You have more functional, more diverse structures if you bring all of the tools of chemistry into DNA,’ she says. Adding synthetic molecules has the bonus that it cuts down on the amount of DNA required. DNA is not cheap to buy; an origami can cost several hundred pounds to construct. 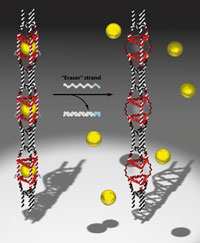
© GRAHAM HAMBLIN
‘We can make any prismatic shape that we want,’ says Sleiman. ‘You can assemble these prismatic cages in minutes at room temperature at 100% yield.’ The polygons are made from single-stranded DNA, with each DNA building block containing a six carbon long alkyl ‘spacer’ to reduce electrostatic and steric crowding.
Sleiman has also made DNA cages containing transition metals. ‘I have always been interested in how to organise metals with DNA because then you can make catalytic factories or sensors or you can make artificial photosynthesis systems,’ she says. Like metallo-supramolecular cages, the metal in metal-DNA cages can be used for redox, photochemical, magnetic and catalytic control over the guest molecules locked inside.
However, making such cages is tricky. Many metals randomly bind to DNA, even cleaving it in some cases. The key is to find the right ligand for the structure. In 2009 Sleiman reported using DNA cages with ligands at each of the vertices that act as pockets for transition metals.
Her group has also made DNA nanotubes which contain guest molecules held in place by DNA strands slightly longer than those in the rest of the structure. Bringing in new DNA strands – ‘eraser’ strands – that are fully complementary to these longer strands peels them away from the nanotubes, freeing the guest molecules.
Inside cells in the body, say in cancer cells, matching strands of mRNA could theoretically trigger the nanotubes to release a drug cargo while non-matching sequences would leave the cargo entrapped. Or the nanotubes could be used as mRNA sensors. ‘Yet one of the big hurdles in the field of DNA therapeutics is the fact that the DNA molecule doesn’t go into cells or if it does, it is only to a limited extent,’ explains Sleiman. She suggests that playing around with the shapes of DNA structures might open a route into cells.
Sleiman is unable to reveal too much about her group’s work to transfer their DNA structures into cells, which is yet to be published. Suffice to say that she is excited about the results.
Turberfield has also had some success in this area. In 2011, his group was able to treat human kidney cells with fluorescently labelled DNA tetrahedra. The cages made their way into the cytoplasm of the cells, remaining ‘substantially intact’ for at least two days. ‘This is the first step towards the use of engineered DNA nanostructures to deliver and control the activity of cargoes within cells,’ he says.
Single molecules

Meanwhile, Rothemun’s origami has a tremendous and growing number of applications, from boards on which to build nanoelectronics to analytical tools. Kurt Gothelf is director of the Centre for DNA Nanotechnology at Aarhus University in Denmark. His team has been studying single molecule chemical reactions on DNA origami and monitoring them using atomic force microscopy (AFM). ‘DNA origami gives you a unique platform for single molecule studies and for studies of distance dependent chemical and biochemical phenomena,’ he says.
Gothelf’s team has used the origami to study the chemistry of singlet oxygen (1O2) which is involved in cell death and many other biological processes, yet is poorly understood. The researchers attach a single photosensitiser molecule (which when irradiated generates singlet oxygen) to a staple strand in the centre of the DNA origami. Also tethered to the origami surface are DNA strands with sections that are susceptible to cleavage by singlet oxygen. On the end of each of these strands is the small molecule biotin. A short period of time after irradiation, the bacterial protein streptavidin is added. Streptavidin has an extremely high affinity for biotin, and therefore attaches itself to the biotin on the staple strands that are untouched by the oxygen. Streptavidin can be visualised using AFM, meaning the researchers can see the intact staple strands, and therefore know which have been cleaved. This technique allows the singlet oxygen’s trail of destruction to be clearly traced.
Gothelf also hopes to make macromolecules by chemical reactions between functional groups in fixed positions on the origami. The idea is that chemical selectivity is determined by the position and orientation of the molecule on the DNA scaffold rather than having to use a series of protecting groups.
[box title=”Making good sense” color=”#dd9933″]At the University of Birmingham, UK, Jim Tucker’s group is developing DNA probes to detect DNA base variations and modifications in the human genome. ‘We make DNA by chemical synthesis in the lab and attach to these DNA strands a probe molecule – a tag [anthracene] – that enables the DNA to be luminescent,’ says Tucker. When the tagged strand interacts with its target to form a double strand, the DNA bases near to the tag quench anthracene luminescence to varying extents, giving a change in emission. The probes can be designed to target single nucleotide polymorphisms (SNPs) in the human genome that can point to an increased risk of developing a disease. Working with Birmingham’s School of Cancer Sciences, the Tucker group has recently developed a sensor for SNPs in a gene that is dysregulated in prostate cancer. The approach differs from many commercial SNP assays, which tend to work through differences in DNA hybridisation efficiency. ‘One challenge in this area is to develop methods which do not rely on the polymerase chain reaction (PCR), for example when monitoring in a cellular environment,’ says Tucker. His team is also developing methods for detecting base modifications in the genome, for example epigenetic effects such as cytosine methylation. Additionally, his group is working on using redox-active tags to create electrochemical DNA sensors. ‘There is quite a lot of interest in electrochemical sensing of DNA. The sensitivity could be as good as that of a luminescent device and the device may be cheaper and more portable,’ says Tucker. [/box]
No help needed
DNA nanodevices that run by themselves without external control hold an obvious appeal for researchers, especially those hoping one day to create a synthetic version of living cells. ‘It is impractical to make molecular motors and operate them from the outside – you need autonomous motion,’ explains Munich’s Simmel. He has been working on ways to drive DNA nanodevices autonomously by coupling them to a complex biochemical process linked to transcription. The system is based on two enzymes used for RNA production and degradation which influence each other by mutual activation and inhibition, creating a kind of oscillating ‘molecular clock’. The clock has been used to control a variety of biomolecular processes including DNA tweezers.
The researchers are also putting these complex systems into micrometre-sized droplets to see what effect the size restriction has. ‘We expect changes in genetics in these little compartments because of the finite size and possibly because of crowding of macromolecules,’ says Simmel.
Spider sense
DNA origami has also paved the way for new generations of basic molecular robots – molecules that can travel. In 2010, a group of US scientists famously made a ‘molecular spider’ which walked along a track of single-stranded DNA tapered to an origami surface. Three of the legs were made from enzymatic DNA which binds to and cuts the DNA strands attached to the origami. When a leg becomes free, after cutting, it explores the origami surface to find another DNA strand to bind to. By repeating this binding and cutting process, the robot followed the track for about 50 steps.
Hao Yan, one of the spider team from the University of Arizona, US, suggests that it may be possible for a spider to pick up a reactant along the way and react it at a specific location to make a ‘molecular factory’. ‘Of course, making tiny amounts of chemicals may not be that practical for applications,’ he adds. He can instead envisage the spider walking along a cell membrane surface and interacting with different receptors, perhaps as a disease treatment.
Oxford’s Turberfield is also working on ‘molecular systems which will perform a series of reactions on a controlled sequence autonomously’. With researchers from Kyoto University, Japan, led by Hiroshi Sugiyama, his team built a 100nm long DNA track and watched a DNA motor move along it using real-time AFM. Adding multiple ‘work stations’ along the track could lead to ‘molecular assembly lines’.

A DNA-based molecular spider has taken 50 steps along a DNA track © PAUL MICHELOTTI
Such assembly lines are inspired by the way that ribosomes in the cell make peptides by moving along mRNA to produce a corresponding chain of amino acids. The DNA track will effectively do the job of the mRNA.
Electronics
Eugen Stulz, a chemist at the University of Southampton, UK, is studying the electronic properties of DNA containing artificial nucleobases. He ‘misuses’ the DNA platform by attaching different functionalities to nucleobases and building them into DNA. With their photoelectronic properties, porphyrins fit the bill perfectly. Stulz’s team has incorporated the molecules into both strands of the DNA duplex. The porphyrins are highly hydrophobic and stack nicely. ‘When the two DNA strands come together the porphyrins act like a zipper, forming a stable arrangement,’ he explains. His team is focusing on the electronic properties of the structures. ‘We are trying to see whether the porphyrin-DNA can function as electronic wires,’ he says.
‘Now that people can see what beautiful structures you can make, many more people are coming into the field,’ says Simmel. ‘Many people feel the potential.’
Ned Seeman, who founded the field of DNA nanotechnology three decades ago, is delighted by its recent exponential growth. ‘For 20 years we were the only laboratory doing this. In the past decade many laboratories have joined us. We no longer have to have all the ideas and we no longer have to make all of the mistakes,’ he jokes. ‘It’s all a lot of fun and it’s very gratifying and rewarding to see so many people bring their own special slant to the field.’
References
S Woo and P W K Rothemund, Nature Chem., 2011, 3, 620 (DOI: 10.1038/nchem.1070)
C K McLaughlin et al, Chem. Commun., 2011, 47, 8925 (DOI: 10.1039/c1cc11726b)
S F J Wickham et al, Nature Nanotech., 2011, 6, 166 (DOI: 10.1038/nnano.2010.284)
S Helmig et al, ACS Nano, 2010, 4, 7475 (DOI: 10.1021/nn102701f)
K Lund et al, Nature, 2010, 465, 206 (DOI: 10.1038/nature09012)
Chemical Society Reviews themed issue on ‘Advances in DNA-based nanotechnology’: Chem. Soc. Rev., 2011, 40, 5623
J-L H A Duprey et al, Chem. Commun., 2011, 47, 6629 (DOI: 10.1039/c1cc11205h)
Story Source:
The above story is based on materials provided by Royal Society of Chemistry, Emma Davies.