Credit: Arizona State University
For more than a decade, scientist Stephen Albert Johnston and his team at Arizona State University’s Biodesign Institute have pooled their energies into an often scoffed-at, high-risk, high-reward goal in medicine: to develop a universal vaccine to prevent cancer.
The mindset is simple, according to Johnston. Treat cancer just like an infectious disease. So, when his team looked deep within tumors, their research gold was discovering 200,000 cancer neoantigens, the components of cancer vaccines, that had been missed by others. They also found that enough of these neoantigens occurred repeatedly in different tumors that it might be possible to make one vaccine for all tumors.
Now, in a new paper published in Scientific Reports, his research team has demonstrated the first experimental proof-of-concept bearing the fruit of their labors.
Their tour-de-force study examined mutations in more than 50 cancer cell lines, and 85 tissue samples from Mayo Clinic Arizona cancer patients, as well as the blood from patients from five different late stage cancer types: lung, breast, brain, gastric and pancreatic cancers.
They have found a common, new source of tumor mutations that could offer three levels of therapy with a cancer vaccine: 1) a broadly protective, or pan-cancer vaccine 2) cancer-type specific vaccines (e.g. breast vs. pancreatic), 3) personalized cancer vaccines based on mutations unique to an individual.
To discover the neoantigens in tumors, Johnston’s team developed a new type of chip. They made chips that presented all of the 200,000 possible neoantigens, allowing them to simply screen for the antibodies in the blood patients raised against them. This is much simpler than the common practice which involves obtaining DNA from the tumor and sequencing it, a starting point for “personal cancer vaccines’ that many companies are now pursuing.
“Personal cancer vaccines are complicated and expensive,” said Johnston. “Also, only about 40% of tumors have enough mutations in the DNA to make a vaccine from. We discovered that even ‘cold tumors’ at the DNA level make lots of mistakes at the RNA level. And the mistakes we focus on are frameshift peptides which are much more immunogenic than the point mutations used in personal cancer vaccines. Most importantly, we can make off-the shelf vaccines for therapeutic or even preventative vaccines which will be much less expensive.”
They report mouse studies showing these new antigens are protective in both breast cancer and melanoma models. Recently, they used this as the basis to begin a large clinical trial in dogs of a prophylactic vaccine designed to be a broadly protective, pan-cancer vaccine. Johnston has also formed the spinout, Calviri, Inc., for further cancer vaccine development and eventually, the first human clinical trials.
A new source for cancer vaccines
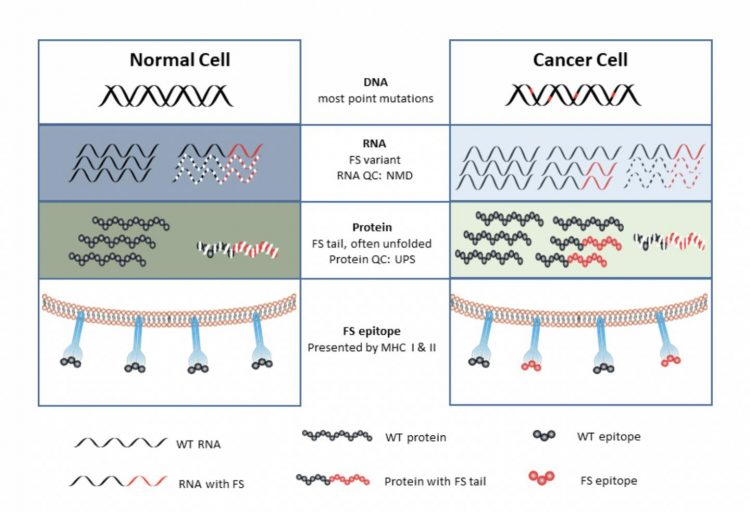
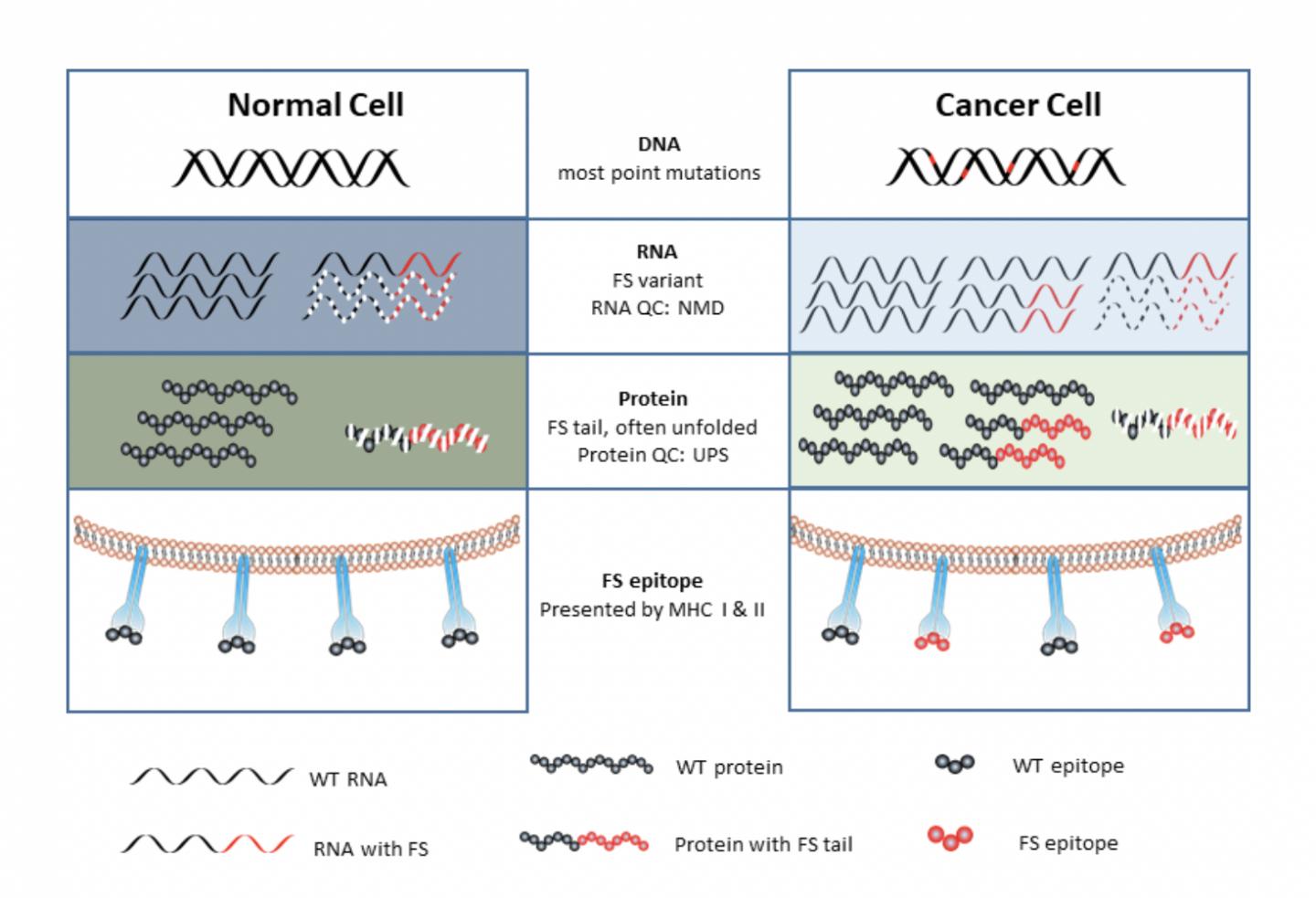
What all of the cancer tumor mutations have in common is making neoantigens, or small protein fragments called peptides made inside a cancer cell that the host’s immune system has never seen before. The aberrant peptides —-only found in cancer cells—-can prime the immune system in a vaccine.
Surprisingly, the source of these neoantigens was found not at the DNA level but rather, errors in the RNA of tumors.
These errors involve changing the reading of the coding for proteins. An analogy is that if you drop the “w” from saw the rat, it will read sat her at.
Similarly, Johnston’s cancer vaccine strategy assumed that there is a general increase in error rates happening when a cell speed reads the instructions in our bodies’ book of DNA code. This cellular information flows from DNA to RNA to proteins, which go on to make all of the things our bodies do, in sickness and in health.
“In a cancer cell, it turns out that all levels of information transfer from DNA to RNA to protein become more error prone,” said Johnston. “We proposed that these mistakes made in cancer cells may also be the source to make a cancer vaccine.”
In one class of alterations, a “frameshift” or “splicing” error occurs when the DNA information in a gene is wrongly processed when making RNA, in a basic cellular process called RNA transcription and splicing.
For the most part, these alterations can be managed and cleaned up by the cell’s intracellular trashcan, never getting exposed to the immune system. By as cancer progresses, as a result of the increase in errors, the protein trash piles up more quickly, overwhelming the cell, and the aberrant proteins are exposed and recognized by the immune cell.
“These overwhelm the quality control systems of a cell, producing mistakes in RNA and proteins that are released from the cancer cell, and the immune system can respond to,” said Johnston.
A vast array
To quickly identify frameshift and splicing mutations, Johnston’s research team designed an array to detect all possible predicted frameshift peptides that any tumor cell could potentially produce. They custom-build this frameshift array, which ended up containing almost 400,000 frameshift peptides, and screened these against the blood samples of cancer patients (and healthy samples as a control) to look for those peptides that had the most reactive antibodies on the array
They found in all five cancer types, with the exception of glioblastoma, the samples had significant peptides reacting with antibodies in the cancer patients than controls.
Within the same cancer type, individuals showed three major patterns:
- 1) The vast majority of the frameshift peptides (69-80%) were personal, or unique for that individual
2) 16-19% of the positive peptides were shared between two samples within the same cancer type
3) 1.5-6.9% were shared between 3 or more (with gastric cancer having the highest of 6.9%)
Strikingly, one of the hardest to treat cancers, glioblastoma, had the greatest potential for personalized vaccines. Of the 17 glioblastoma patient samples studied, each patient had 5800 frameshift peptides, and of these, 4,500 were unique for that patient.
They also wanted to see how the frameshift mutations compared with early and late stage cancers. They compared 20,000 peptides that they identified in late stage and stage 1 pancreatic cancer, and showed that there was little overlap between them, implying the vaccine for early stage cancer would be different than that for late stage cancer.
The magic bullets
All of the frameshift alterations, identification and screening experiments allowed them to choose the top vaccine candidates. These were tested in several mouse studies in a variety of cancer prevention and therapy challenges.
Johnston group has also pioneered genetic immunization and the gene gun, which they used to shoot gold nanoparticles containing the most promising vaccines. In a typical experiment, six-week-old mice received one genetic immunization in the pinna of the ear. After four weeks, they were challenged with cancer-causing cells, then, twice received booster shots two days apart.
They found that these vaccines could all significant delay or even prevent tumor growth or prevent progression. Most importantly, in the mouse vaccine challenges, they found pooling multiple frameshift peptides produced a significant additive increase in delaying tumor growth, and made for a more effective vaccine.
Top guns
From their screening and analyses of the different cancer samples, and the mouse cancer vaccine challenges, they now have a “top 100” peptide list for each of the five human cancers. They have steadily moved from concept to discovery to mice studies to the first-ever dog cancer vaccine trial, funded by the Open Philanthropy Project. If that is successful, Johnston is eager to move to the first human clinical trials.
Johnston said they have the technology to make the human vaccine right now, but even optimistically it would be five to 10 years before human use.
“This is probably the only approach to a broadly preventative cancer vaccine, so we feel we have to try it,” said Johnston. “The implications of success would be quite large — for dogs and people.”
###
The work was the culmination of years of support, including early, high-risk grants from the Department of Defense and the Keck Foundation. More than 20 members of Johnston’s team from the Biodesign Center for Innovations in Medicine performed the work, including Phillip Stafford, Tsukasa Oyama and Bao-Xi Qu for early foundational research and statistical analyses.
Media Contact
Joseph Caspermeyer
[email protected]
Related Journal Article
http://dx.