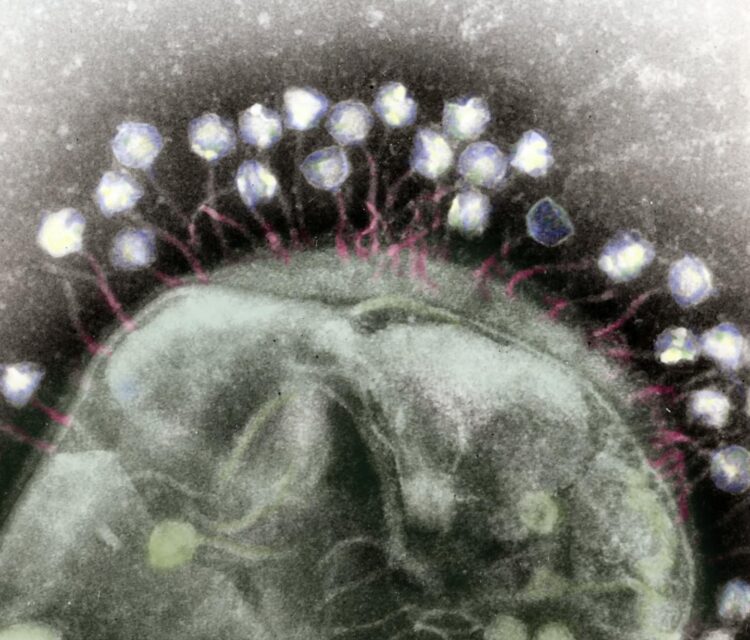
A new genetic approach can accelerate the study of phage-microbe interactions with implications for health, agriculture, and climate
Credit: Wikimedia Commons
Scientists are continually searching for new and improved ways to deal with bacteria, be it to eliminate disease-causing strains or to modify potentially beneficial strains. And despite the numerous clever drugs and genetic engineering tools humans have invented for these tasks, those approaches can seem clumsy when compared to the finely tuned attacks waged by phages – the viruses that infect bacteria.
Phages, like other parasites, are continually evolving ways to target and exploit their specific host bacterial strain, and in turn, the bacteria are continually evolving means to evade the phages. These perpetual battles for survival yield incredibly diverse molecular arsenals that researchers are itching to study, yet doing so can be tedious and labor-intensive.
To gain insight into these defensive strategies, a team led by Berkeley Lab scientists has just developed an efficient and inexpensive new method. As reported in PLOS Biology, the team showed that a combination of three techniques can reveal which bacterial receptors phages exploit to infect the cell, as well as what cellular mechanisms the bacteria use to respond to a phage infection.
“Despite nearly a century of molecular work, the underlying mechanisms of phage-host interactions are only known for a few pairs, where the host is a well-studied model organism that can be cultured in a lab,” said corresponding author Vivek Mutalik, a research scientist in Berkeley Lab’s Environmental Genomics and Systems Biology (EGSB) Division. “However, phages represent the most abundant biological entities on Earth, and due to their impact on bacteria, they are key drivers of environmental nutrient cycles, agricultural output, and human and animal health. It has become imperative to gain more foundational knowledge of these interactions in order to better understand the planet’s microbiomes and to develop new medicines, such as bacteria-based vaccines or phage cocktails to treat antibiotic-resistant infections.”
Shining a light on “dark matter”
The team’s three-pronged approach, called barcoded loss-of-function and gain-of-function libraries, uses the established technique of creating gene deletions and also increasing gene expression to identify which genes the bacteria use to evade the phages. This information also tells the scientists which receptors the phages are targeting without having to analyze the phages’ genomes. (However, the scientists do plan to adapt the technique for use on viruses in the future, to learn even more about their function.)
Mutalik and his colleagues tested their method on two strains of E. coli that are known to be targeted by 14 genetically diverse phages. Their results confirmed that the method works smoothly by speedily revealing the same suite of phage receptors that had been previously identified through decades of research, and also provided new hits that were missed in earlier studies.
According to Mutalik, the approach can also be scaled-up to simultaneously evaluate phage relationships for hundreds of bacteria sampled from diverse environments. This will make it much easier for scientists to study the planet’s biological “dark matter,” which refers to the unculturable and therefore poorly understood microorganisms that abound in many environments. In fact, it is estimated that 99% of all living microorganisms can’t be cultured in a lab.
The team’s approach also represents an opportunity to standardize genetic resources used in phage research, which has always been an ad-hoc and highly variable process, and create sharable reagents and datasets.
“The role of phages is a huge ‘known-unknown,’ as we know there are phages everywhere, but hardly know anything more. For example, we understand less than 10% of the genes encoded in previously sequenced phage genomes,” said Mutalik. “Now that we finally have a streamlined tool to look at phages, there are many exciting questions we can start to answer and an opportunity to make a difference in the world.”
###
This work was led by Mutalik and fellow EGSB scientists Adam Arkin and Adam Deutschbauer at Berkeley Lab, in collaboration with researchers at UC Berkeley and Evergreen State College. The research was funded by the Microbiology program of the Innovative Genomics Institute and by ENIGMA (https:/
Founded in 1931 on the belief that the biggest scientific challenges are best addressed by teams, Lawrence Berkeley National Laboratory and its scientists have been recognized with 14 Nobel Prizes. Today, Berkeley Lab researchers develop sustainable energy and environmental solutions, create useful new materials, advance the frontiers of computing, and probe the mysteries of life, matter, and the universe. Scientists from around the world rely on the Lab’s facilities for their own discovery science. Berkeley Lab is a multiprogram national laboratory, managed by the University of California for the U.S. Department of Energy’s Office of Science.
DOE’s Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit energy.gov/science.
Media Contact
Aliyah Kovner
[email protected]
Original Source
https:/
Related Journal Article
http://dx.
https://scienmag.com/cataloging-natures-hidden-arsenal-viruses-that-infect-bacteria/