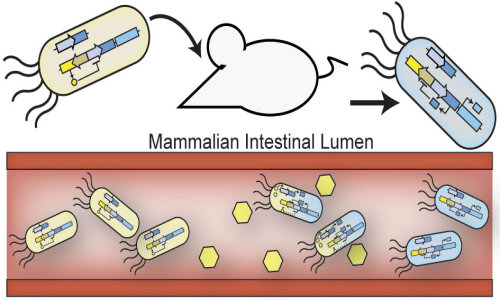
It’s a jungle in there. In the tightly woven ecosystem of the human gut, trillions of bacteria compete with each other on a daily basis while they sense and react to signals from the immune system, ingested food, and other bacteria.
Problems arise when bad gut bugs overtake friendly ones, or when the immune system is thrown off balance, as in Crohn’s disease, celiac disease, and colorectal cancer. Doctors have struggled to diagnose these conditions early and accurately. But now a new engineered strain of E. coli bacteria could deliver status updates from this complex landscape to help keep gastrointestinal diseases at bay.
The new strain non-destructively detected and recorded an environmental signal in the mouse gut, and remembered what it “saw.” The advance, reported in the Proceedings of the National Academy of Sciences, could lead to a radically new screening tool for human gut health.
Inspired by Nature, the team engineered E. coli to sense, record and remember an environmental signal in the gut — and also demonstrated that they can survive and function within complex environments such as the mammalian gut. This work lays the foundation for the use of engineered probiotic bacteria that serve as living diagnostics. In this schematic engineered probiotic E. coli have colonized the mammalian intestine and “remember” exposure to an environmental signal, which is indicated by the cells turning blue in color. Credit: Jonathan Kotula, Harvard’s Wyss Institute and Harvard Medical School
The key to turning E. coli into gut reporters was to insert a well-known genetic switch that flips when it senses a specific environmental cue. This switch confers on the cells the ability to “remember” what they sense for up to a week – long enough for scientists to recover fecal samples and test whether the switch has flipped.
“This achievement paves the way toward living monitors programmed using synthetic gene circuits,” said Wyss Institute Core Faculty member Pamela Silver, Ph.D., senior author on the study who is also the Elliott T. and Onie H. Adams Professor of Biochemistry and Systems Biology at Harvard Medical School (HMS). Silver’s team included James Collins, Ph.D., who is also a Wyss Core Faculty member and professor of bioengineering at Boston University, as well as other collaborators from the Wyss Institute, Harvard Medical School and Boston University. “It could lead to new diagnostics for all sorts of complex environments.”
The approach Silver’s team took runs counter to the prevailing dogma in synthetic biology, which is to design genetic systems that drive cell behavior from scratch, said Wyss Institute Senior Staff Scientist Jeff Way, Ph.D., a coauthor on the paper. On the other hand, “Nature has a tried-and-true blueprint for memory systems if you know where to look,” Way said. “Why not just accept Nature as it is, and develop the system from there?”
The genetic switch the team inserted in E. coli came from lambda phage, a virus that commonly attacks this bacterium.
After invading E. coli, lambda typically lays low, living in a stealth mode called lysogeny in which its DNA simply hangs out in the E. coli’s genome. But when the bacterium’s DNA is damaged – and only then – the switch flips, instructing the virus to enter a mode called lysis in which it multiplies inside the cell and breaks through its membrane in a kind of microbial explosion.
“This is a very stable system in Nature,” said lead author Jonathan Kotula, Ph.D., a Postdoctoral Fellow at HMS who is also affiliated with the Wyss Institute. “We knew the lambda switch would be a great candidate for the memory element, and we simply tweaked it to meet our needs.”
The cells with the engineered lambda switch would not become lytic under any conditions. Kotula and the rest of Silver’s team used standard molecular genetic tools to rig the switch such that it turned on only in the presence of an inactive form of the antibiotic tetracycline.
In laboratory experiments, the switch turned on within a few hours of exposure to the antibiotic– and stayed in this ‘ON’ state inside E. coli for a week or more, even as the bacteria grew and divided. In short, the cells “remembered” that they had seen that molecule in the gut.
“It was truly shocking how cleanly the experiments worked,” said Jordan Kerns, Ph.D., a Wyss Institute Postdoctoral Fellow.
But to function as a living diagnostic, the engineered E. coli also had to survive their trip through the gut intact, which meant they had to compete effectively against rival gut microbes.
The engineered strain worked fine in laboratory experiments, but gradually disappeared when the team introduced it into the gut of the mouse itself. It turned out that it had been outcompeted by the animal’s native gut bacteria. The team did not fret in the face of this result because they knew that the classical strain of E. coli they used had lived only in the laboratory since the 1940s – losing its ability to compete in the real world, particularly in an environment as challenging as the mammalian gut.
They tackled the problem by isolating a native strain of E. coli from the mouse gut, then engineering its genome to incorporate the switch. The switch in the cells flipped within hours, as it had before, and the cells “remembered” for about a week that they had seen the antibiotic in the gut, Kotula said. Moreover, the population stabilized within the gut, holding its own in the presence of other bacteria.
The team envisions a day when a doctor would give a patient a strain of engineered bacteria as a diagnostic, much as they give a probiotic today. The strain would be rigged to monitor the gut for any number of conditions from inflammation to disease markers. At a follow-up visit, the patient would submit a stool sample, and medical technicians would collect E. coli from the sample and analyze it. Only if the switch (or switches) were on would the doctor perform more invasive tests such as an endoscopy or a colonoscopy.
For now the team is focusing on genetically tweaking the memory element of their system so that the cells remember for even longer periods of time, and engineering it so the switch flips when it senses other chemical signatures as well, such as those of cancer or parasites. In the longer term, their engineered bacteria could sense a disease state and work with other engineered genetic circuits that can produce a specific drug on command, thus producing a dynamic therapy.
“Our increasing appreciation of the role of the microbiome in health and disease is transforming the entire medical field. The concept of using the power of synthetic biology to harness microbes that live in our gut to develop living diagnostic and therapeutic devices is a harbinger of things to come, and Pam’s work provides the first proof-of-principle that this is a viable and exciting path to pursue,” said Wyss Institute Founding Director Don Ingber, Ph.D., M.D.
Story Source:
The above story is based on materials provided by Harvard University.