Without enzymes, an organism would not be able to survive. It is these biocatalysts that facilitate a whole range of chemical reactions, producing the building blocks of the cells. Enzymes are also used widely in biotechnology and in our households, where they are used in detergents, for example.
Credit: HHU / Swastik Mishra
Without enzymes, an organism would not be able to survive. It is these biocatalysts that facilitate a whole range of chemical reactions, producing the building blocks of the cells. Enzymes are also used widely in biotechnology and in our households, where they are used in detergents, for example.
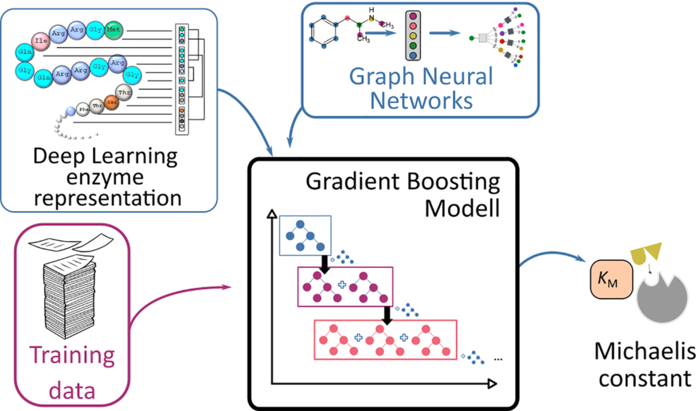
To describe metabolic processes facilitated by enzymes, scientists refer to what is known as the Michaelis-Menten equation. The equation describes the rate of an enzymatic reaction depending on the concentration of the substrate – which is transformed into the end products during the reaction. A central factor in this equation is the ‘Michaelis constant’, which characterises the enzyme’s affinity for its substrate.
It takes a great deal of time and effort to measure this constant in a lab. As a result, experimental estimates of these constants exist for only a minority of enzymes. A team of researchers from the HHU Institute of Computational Cell Biology and Chalmers University of Technology in Stockholm has now chosen a different approach to predict the Michaelis constants from the structures of the substrates and enzymes using AI.
They applied their approach, based on deep learning methods, to 47 model organisms ranging from bacteria to plants and humans. Because this approach requires training data, the researchers used known data from almost 10,000 enzyme-substrate combinations. They tested the results using Michaelis constants that had not been used for the learning process.
Prof. Lercher had this to say about the quality of the results: “Using the independent test data, we were able to demonstrate that the process can predict Michaelis constants with an accuracy similar to the differences between experimental values from different laboratories. It is now possible for computers to estimate a new Michaelis constant in just a few seconds without the need for an experiment.”
The sudden availability of Michaelis constants for all enzymes of model organisms opens up new paths for metabolic computer modelling, as highlighted by the journal PLOS Biology in an accompanying article.
Original publication
Alexander Kroll, Martin K. M. Engqvist, David Heckmann, Martin Lercher, Deep learning allows genome-scale prediction of Michaelis constants from structural features, PLOS Biology (2021).
DOI: 10.1371/journal.pbio.3001402
Journal
PLoS Biology
DOI
10.1371/journal.pbio.3001402
Article Title
Deep learning allows genome-scale prediction of Michaelis constants from structural features
Article Publication Date
19-Oct-2021




