New optical sensing platform can detect genomic cancer biomarkers directly in patient tissues
Credit: Tuan Vo-Dinh, Duke University
Biomedical engineers at Duke University have engineered a method for simultaneously detecting the presence of multiple specific microRNAs in RNA extracted from tissue samples without the need for labeling or target amplification. The technique could be used to identify early biomarkers of cancer and other diseases without the need for the elaborate, time-consuming, expensive processes and special laboratory equipment required by current technologies.
The results appeared online on May 4 in the journal Analyst.
“The general research focus in my lab has been on the early detection of diseases in people before they even know they’re sick,” said Tuan Vo-Dinh, director of the Fitzpatrick Institute for Photonics and the R. Eugene and Susie E. Goodson Distinguished Professor of Biomedical Engineering at Duke. “And to do that, you need to be able to go upstream, at the genomic level, to look at biomarkers like microRNA.”
MicroRNAs are short RNA molecules that bind to messenger RNA and stop them from delivering their instructions to the body’s protein-producing machines. This could effectively silence certain sections of DNA or regulate gene expression, thus altering the behaviors of certain biological functions. More than 2000 microRNAs have been discovered in humans that affect development, differentiation, growth and metabolism.
As researchers have discovered and learned more about these tiny genetic packages, many microRNAs have been linked to the misregulation of biological functions, resulting in diseases ranging from brain tumors to Alzheimer’s. These discoveries have led to an increasing interest in using microRNAs as disease biomarkers and therapeutic targets. Due to the very small amounts of miRNAs present in bodily samples, traditional methods of studying them require genetic-amplification processes such as quantitative reverse transcription PCR (qRT-PCR) and RNA sequencing.
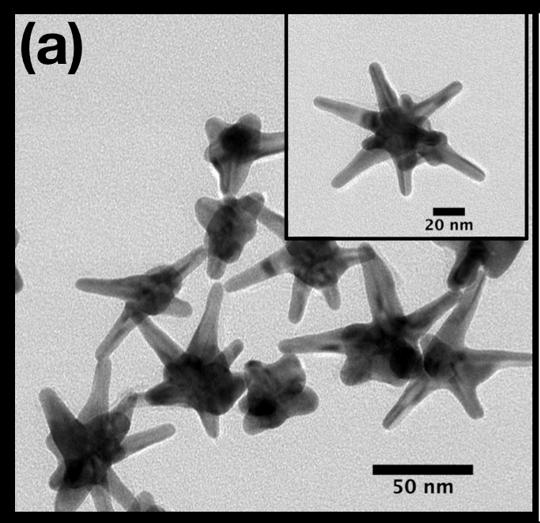
While these technologies perform admirably in well-equipped laboratories and research studies that can take months or years, they aren’t as well-suited for fast diagnostic results at the clinic or out in the field. To try to bridge this gap in applicability, Vo-Dinh and his colleagues are turning to silver-plated gold nanostars.
“Gold nanostars have multiple spikes that can act as lighting rods for enhancing electromagnetic waves, which is a unique feature of the particle’s shape,” said Vo-Dinh, who also holds a faculty appointment in Duke chemistry. “Our tiny nanosensors, called ‘inverse molecular sentinels,’ take advantage of this ability to create clear signals of the presence of multiple microRNAs.”
While the name is a mouthful, the basic idea of the nanosensor design is to get a label molecule to move very close to the star’s spikes when a specific stretch of target RNA is recognized and captured. When a laser is then shined on the triggered sensor, the lightning rod effect of the nanostar tips causes the label molecule to shine extremely brightly, signaling the capture of the target RNA.
The researchers set this trigger by tethering a label molecule to one of the nanostar’s points with a stretch of DNA. Although it’s built to curl in on itself in a loop, the DNA is held open by an RNA “spacer” that is tailored to bind with the target microRNA being tested for. When that microRNA comes by, it sticks to and removes the spacer, allowing the DNA to curl in on itself in a loop and bring the label molecule in close contact with the nanostar.
Under laser excitation, that label emits a light called a Raman signal, which is generally very weak. But the shape of the nanostars–and a coupling effect of separate reactions caused by the gold nanostars and silver coating–amplifies Raman signals several million-folds, making them easier to detect.
“The Raman signals of label molecules exhibit sharp peaks with very specific colors like spectral fingerprints that make them easily distinguished from one another when detected,” said Vo-Dinh. “Thus we can actually design different sensors for different microRNAs on nanostars, each with label molecules exhibiting their own specific spectral fingerprints. And because the signal is so strong, we can detect each one of these fingerprints independently of each other.”
In this clinical study, Vo-Dinh and this team collaborated with Katherine Garman, associate professor of medicine, and colleagues at the Duke Cancer Institute to use the new nanosensor platform to demonstrate that they can detect miR-21, a specific microRNA associated with very early stages of esophageal cancer, just as well as other more elaborate state-of-the-art methods. In this case, the use of miR-21 alone is enough to distinguish healthy tissue samples from cancerous samples. For other diseases, however, it might be necessary to detect several other microRNAs to get a reliable diagnosis, which is exactly why the researchers are so excited by the general applicability of their inverse molecular sentinel nanobiosensors.
“Usually three or four genetic biomarkers might be sufficient to get a good diagnosis, and these types of biomarkers can unmistakably identify each disease,” said Vo-Dinh. “That’s why we’re encouraged by just how strong of a signal our nanostars create without the need of time-consuming target amplification. Our method could provide a diagnostic alternative to histopathology and PCR, thus simplifying the testing process for cancer diagnostics.”
For more than three years, Vo-Dinh has worked with his colleagues and Duke’s Office of Licensing and Ventures to patent his nanostar-based biosensors. With that patent recently awarded, the researchers are excited to begin testing the limits of their technology’s abilities and exploring technology transfer possibilities with the private sector.
“Following these encouraging results, we are now very excited to apply this technology to detect colon cancer directly from blood samples in a new NIH-funded project,” said Vo-Dinh. “It’s very challenging to detect early biomarkers of cancer directly in the blood before a tumor even forms, but we have high hopes.”
###
This research was supported by the National Institutes of Health (1R21CA196426) and the National Science Foundation (1106401).
CITATION: “Plasmonic Nanobiosensors for Detection of MicroRNA Cancer Biomarkers in Clinical Samples,” Bridget M. Crawford, Hsin-Neng Wang, Christina Stolarchuk, Richard J. von Furstenberg, Pietro Strobbia, Dadong Zhang, Xiaodi Qin, Kouros Owzar, Katherine S. Garman, Tuan Vo-Dinh. Analyst, May 4, 2020. DOI: 10.1039/D0AN00193G
Media Contact
Ken Kingery
[email protected]
Original Source
https:/
Related Journal Article
http://dx.




