Researchers characterize 3 ways in which the gene FOXA1 mutates to trigger prostate cancer
Credit: University of Michigan Rogel Cancer Center
ANN ARBOR, Michigan — The Fast and the Furious movie franchise meets the Fast N’ Loud television series to define an oncogene that drives 35% of prostate cancers.
A new study from researchers at the University of Michigan Rogel Cancer Center finds that the gene FOXA1 overrides normal biology in three different ways to drive prostate cancer. They refer to the three classes as FAST, FURIOUS, and LOUD to reflect their unique features. The findings are published in Nature.
“It’s quite intriguing and complex biology,” says senior study author Arul M. Chinnaiyan, M.D., Ph.D., director of the Michigan Center for Translational Pathology and S.P. Hicks Endowed Professor of Pathology at Michigan Medicine.
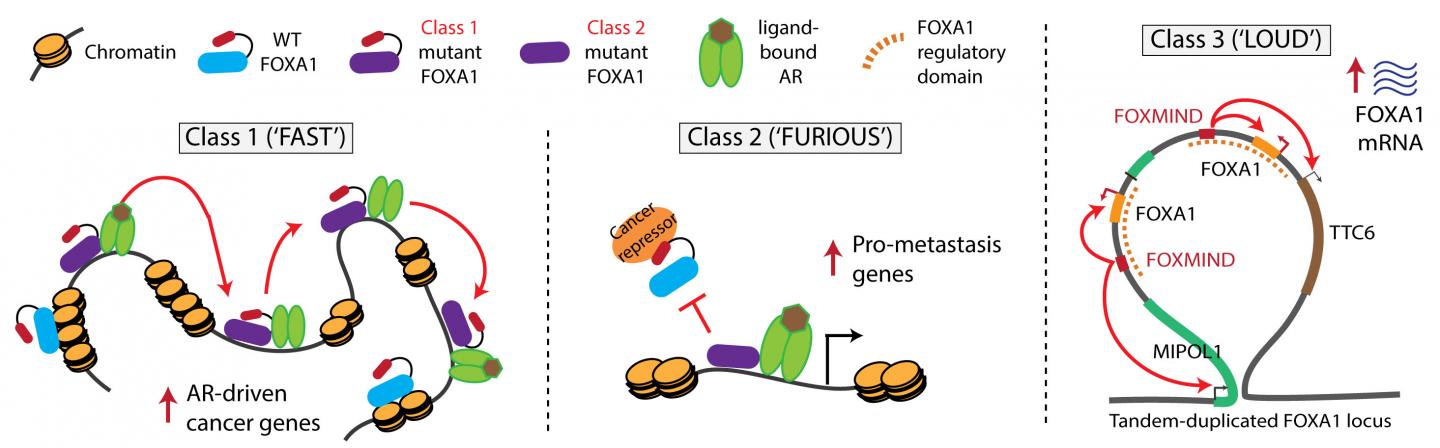
“We found that the same gene can be turned into an oncogene in three different ways,” says Abhijit Parolia, a molecular and cellular pathology graduate student and co-first author on this study. “One moves fast in the nucleus, the second binds to chromatin furiously and the third amplifies itself to be loud. These three alteration classes have different clinical implications for patients.”
Class 1 mutations are FAST. They cause the transcription factor to travel more quickly through the DNA, allowing the partnering androgen receptor to activate expression of cancer-promoting genes. Imagine the driver racing forward at high speed. These mutations are seen in early stage prostate cancer and are likely what triggers the disease.
Class 2 mutations are FURIOUS. The mutation causes a portion of the FOXA1 molecule to be cut off. This truncated molecule binds very strongly to the DNA, preventing normal FOXA1 from binding. These mutations are found in lethal hormone-therapy resistant prostate cancer and promote the cancer’s spread to distant sites. Think of the mutant as furiously binding DNA and dominantly enabling the cancer’s aggressive features.
Class 3 mutations are LOUD. They involve complex rearrangements of the FOXA1 genomic position, creating duplications in which FOXA1 or other oncogenes are overexpressed. In other words, the amplified oncogenes work at top volume to be biologically heard. This can occur in both early stage and metastatic cancer.
Fast and furious mutations are mutually exclusive but loud rearrangements can exist by themselves or mingle with either of the other two.
FOXA1 was previously known to be mutated in prostate cancer, but its biological functions were poorly understood. Scientists were uncertain if FOXA1 was an oncogene that fueled cancer or a tumor suppressor that hit the brakes. The Rogel Cancer Center team now clarified FOXA1’s role as a driver oncogene, in addition to classifying the three novel FOXA1 alterations.
The researchers discovered its increased prevalence by using RNA sequencing data from 1,546 prostate cancer samples from multiple collections, including the Rogel Cancer Center’s Mi-ONCOSEQ program.
“Oncogenes tend to be easier to develop therapies for as you could theoretically block them with targeted medicines,” Chinnaiyan says. “However, FOXA1 is a challenging target because it is a transcription factor, a class of proteins notoriously difficult to inhibit with small molecules. However, scientists are now developing innovative strategies to go after these ‘undruggable’ targets.”
Chinnaiyan says this information can also be used to identify patients with more aggressive disease or begin to understand why patients respond to therapy differently.
The authors also showed that the three classes of FOXA1 alterations are found in breast cancer, presumably impacting the estrogen receptor in a way similar to how it impacts the androgen receptor. FOXA1 alterations are implicated in bladder cancer and some salivary gland cancers as well.
###
Additional authors: Abhijit Parolia, Marcin Cieslik, Shih-Chun Chu, Lanbo Xiao, Takahiro Ouchi, Yuping Zhang, Xiaoju Wang, Pankaj Vats, Xuhong Cao, Sethuramasundaram Pitchiaya, Fengyun Su, Rui Wang, Felix Y. Feng, Yi-Mi Wu, Robert J. Lonigro, Dan R. Robinson
Funding: Prostate Cancer Foundation; National Cancer Institute grants U01 CA214170 and P50 CA186786; Stand Up 2 Cancer-PCF Dream Team grant SU2C-AACR-DT0712; Howard Hughes Medical Institute; A. Alfred Taubman Institute; American Cancer Society; Department of Defense grants W81XWH-17-1-0130, W81XWH-17-1-0224, W81XWH-16-1-0195
Disclosure: None
Reference: Nature, doi: 10.1038/s41586-019-1347-4, published online June 26, 2019
Resources:
University of Michigan Rogel Cancer Center, http://www.
Michigan Health Lab, http://www.
Michigan Medicine Cancer AnswerLine, 800-865-1125
Media Contact
Nicole Fawcett
[email protected]
Related Journal Article
http://dx.




