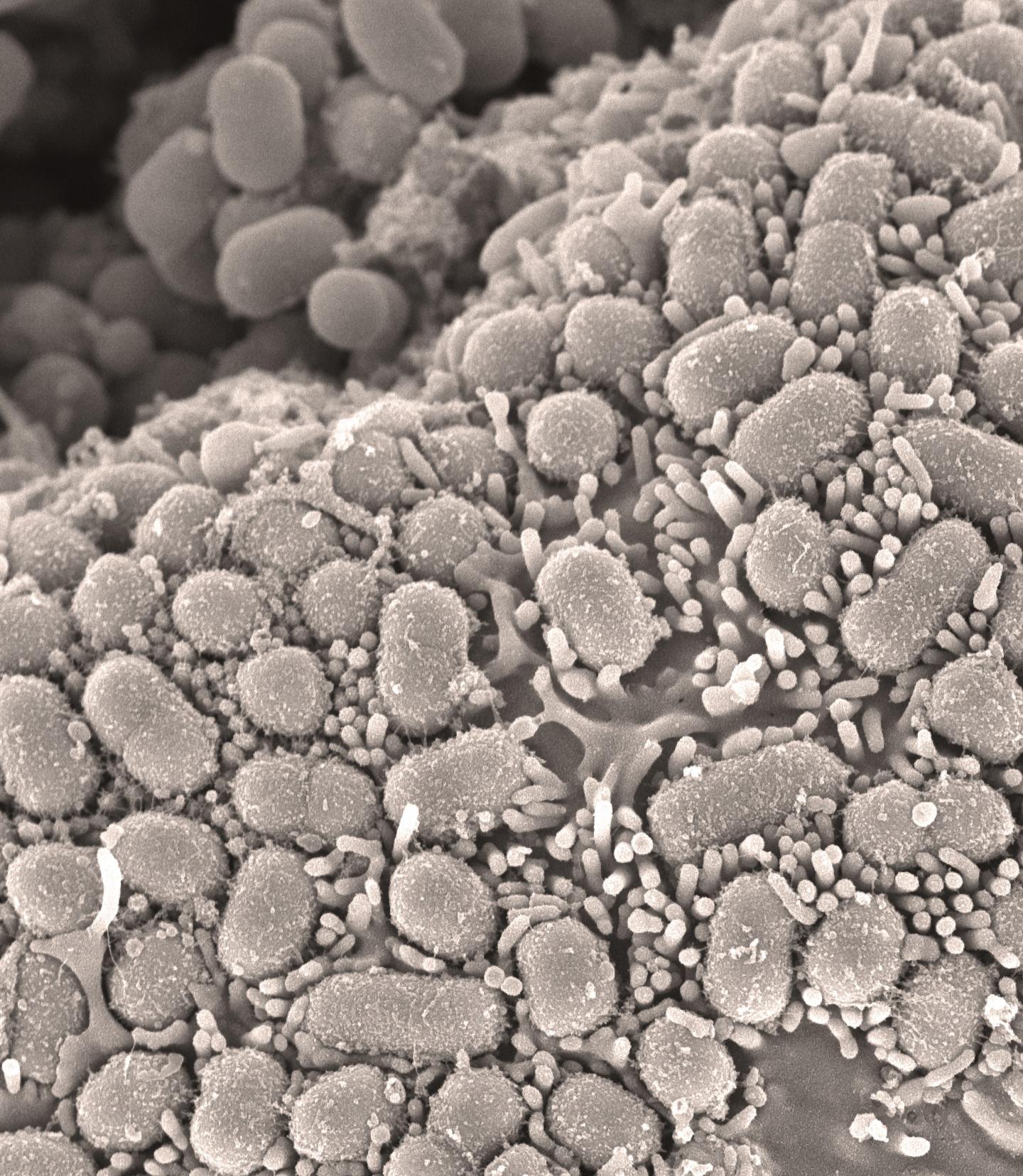
Credit: David Goulding (Wellcome Sanger Institute)
Some MRSA infections could be tackled using widely-available antibiotics, suggests new research from an international collaboration led by scientists at the University of Cambridge and the Wellcome Sanger Institute.
Since the discovery of penicillin, the introduction of antibiotics to treat infections has revolutionised medicine and healthcare, saving millions of lives. However, widespread use (and misuse) of the drugs has led some bacteria to develop resistance, making the medicines less effective. With few new antibiotics in development, antibiotic resistance is widely considered a serious threat to the future of modern medicine, raising the spectre of untreatable infections.
One of the most widely used and clinically important groups of antibiotics is the family that includes penicillin and penicillin derivatives. The first type of penicillin resistance occurred when bacteria acquired an enzyme, known as a beta-lactamase, which destroys penicillin. To overcome this, drug manufacturers developed new derivatives of penicillin, such as methicillin, which were resistant to beta-lactamase.
In the escalating arms race, one particular type of bacteria known as Methicillin-resistant Staphylococcus aureus – MRSA – has developed widespread resistance to this class of drugs. MRSA has become a serious problem in hospital- and community-acquired infections, forcing doctors to turn to alternative antibiotics, or a cocktail of different drugs which are often less effective, and raises concerns that even these drugs will in time become ineffective.
In previous research, a team of researchers in Cambridge identified an isolate of MRSA (a sample grown in culture from a patient’s infection) that showed susceptibility to penicillin in combination with clavulanic acid. Clavulanic acid is a beta-lactamase inhibitor, which prevents the beta-lactamase enzyme destroying penicillin; it is already used as a medicine to treat kidney infections during pregnancy.
In a study published today in Nature Microbiology, a team of scientists from the UK, Denmark, Germany, Portugal, and USA used genome sequencing technology to identify which genes make MRSA susceptible to this combination of drugs. They identified a number of mutations (changes in the DNA sequence) centred around a protein known as a penicillin-binding protein 2a or PBP2a.
PBP2a is crucial to MRSA strains as it enables them to keep growing in the presence of penicillin and other antibiotics derived from penicillin. Two of these mutations reduced PBP2a expression (the amount of PBP2a produced), while two other mutations increased the ability of penicillin to bind to PBP2a in the presence of clavulanic acid. Overall the effect of these mutations means that a combination of penicillin and clavulanic acid could overcome the resistance to penicillin in a proportion of MRSA strains.
The team then looked at whole genome sequences of a diverse collection of MRSA strains and found that a significant number of strains – including USA300 clone, the dominant strain in the United States – contained both mutations that confer susceptibility. This means that one of the most widespread strains of MRSA-causing infections could be treatable by a combination of drugs already licensed for use.
Using this knowledge, the researchers used a combination of the two drugs to successfully treat MRSA infections in moth larvae and then mice. Their next step will be to conduct the further experimental work required for a clinical trial in humans.
Dr Mark Holmes from the Department of Veterinary Medicine at the University of Cambridge, a senior author of the study, says: “MRSA and other antibiotic-resistant infections are a major threat to modern medicine and we urgently need to find new ways to tackle them. Developing new medicines is extremely important, but can be a lengthy and expensive process. Our works suggests that already widely-available medicines could be used to treat one of the world’s major strains of MRSA.”
First author Dr Ewan Harrison, from the Wellcome Sanger Institute and the University of Cambridge, adds: “This study highlights the importance of genomic surveillance – collecting and sequencing representative collections of bacterial strains. By combining the DNA sequencing data generated by genomic surveillance with laboratory testing of the strains against a broad selection of antibiotics, we may find other unexpected chinks in the armour of antibiotic-resistant bacteria that might give us new treatment options.”
The research was funded by the Medical Research Council (MRC), Wellcome and the Department of Health.
Dr Jessica Boname, Head of Antimicrobial Resistance at the MRC, says: “This study demonstrates how a mechanistic understanding of resistance and access to clinical data can be used to find new ways to contain and control infections with existing resources.”
###
Reference
Harrison, EM et al. Genomic identification of cryptic susceptibility to penicillins and β-lactamase inhibitors in methicillin-resistant Staphylococcus aureus. Nature Microbiology; 24 June 2019; DOI: 10.1038/s41564-019-0471-0
Media Contact
Craig Brierley
[email protected]
Related Journal Article
http://dx.