‘Worldwide, breast cancer is the most common cancer in women.’
Oncotarget published “Landscape of somatic mutations in breast cancer: new opportunities for targeted therapies in Saudi Arabian patients” which reported that the association between genetic polymorphisms in tumor suppressor genes and the risk of BCa has been studied in many ethnic populations with conflicting conclusions while Arab females and Saudi Arabian studies are still lacking.
The authors screened a cohort of Saudi BCa patients by NGS using a bespoke gene panel to clarify the genetic landscape of this population, correlating and assessing genetic findings with clinical outcomes.
They identified a total of 263 mutations spanning 51 genes, including several frequently mutated.
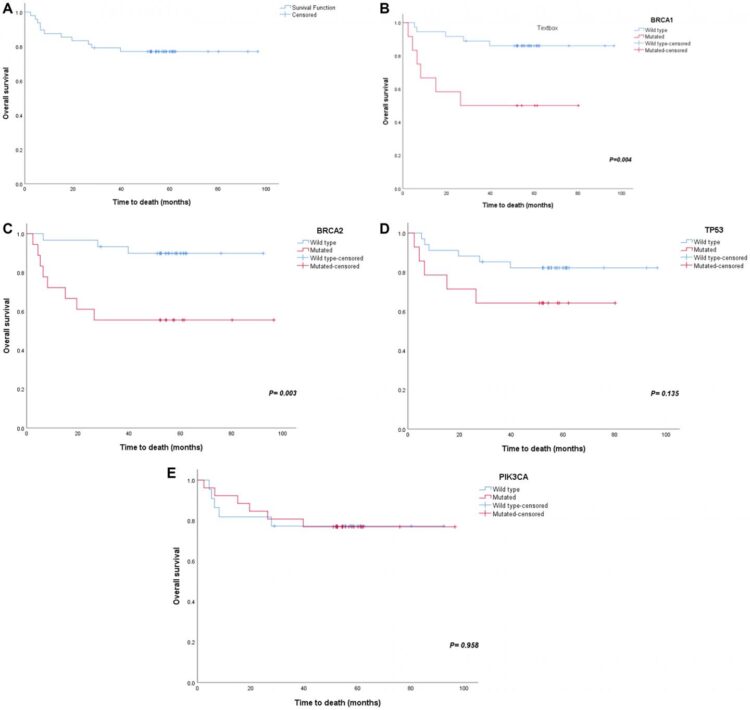
Among the genes analyzed, the highest mutation rates were found in PIK3CA, BRCA2, BRCA1, TP53, MSH2, PMS2, BARD1, MLH1, CDH1, RAD50, MSH6, NF1, in addition to others.
They identified multiple common recurrent variants and previously reported mutations.
They also reveal the landscape of the mutations associated with BCa in Saudi women, highlighting the importance of routine genetic sequencing in implementation of precision therapies in KSA.
Dr. Malak Abedalthagafi from The King Fahad Medical City and King Abdulaziz City for Science and Technology said, “Worldwide, breast cancer is the most common cancer in women.”
“Worldwide, breast cancer is the most common cancer in women.”
A positive ER and/or PR, either positive HER2 or negative HER2, and high levels of Ki67 suggest a luminal B BCa subtype, which makes up less than 20% of all BCa cases and has lower survival rates than luminal A.
The absence of ER and PR expression accompanied by high expression of HER2 and proliferation gene clusters and low expression of luminal and basal clusters, as detected by IHC, suggests a HER2-enriched BCa subtype, which accounts for 10% to 15% of all cases and has a poorer prognosis than luminal cancers.
Three significant pathways govern mammary gland and BCa stem cell development:
Estrogen receptor signaling;
HER2 signaling; and
Canonical Wnt signaling.
In ER signaling, estrogen binds membrane estrogen receptors and triggers a cascade of events that ultimately promote the binding of nuclear estrogen receptors with estrogen response elements.
Other pathways involved in BCa development include cyclin-dependent kinase signaling, notch signaling, sonic hedgehog signaling, breast tumor kinase signaling, and PI3K/AKT/mTOR signaling.
WES also facilitated both the identification of the FANCM gene as a susceptible gene for triple-negative BCa and the association of XCR1, DLL1, TH, ACCS, SPPL3, CCNF and SRL with BCa.
In this study, they screened a cohort of Saudi BCa patients using a cancer-specific gene panel to ascertain the mutation spectrum and explore the possible clinical implications of the identified somatic variants in BCa development.
The Abedalthagafi Research Team concluded in their Oncotarget Research Output that the authors identified somatic mutation variants in Saudi BCa patients; BRCA1, BRCA2, TP53, and PIK3CA were found to be among the most common.
In total, they identified 39 novel mutations that were not reported before and were predicted to be pathogenic.
Their study has pertinent limitations.
Further, their limited sample size, particularly for limited somatic genomics aberrations analyzes, may limit generalizability.
More regional studies are still needed.
###
DOI – https:/
Full text – https:/
Correspondence to – Malak Abedalthagafi – [email protected]
Keywords – breast cancer, PIK3CA, BCa, Saudi Arabia, BRCA
About Oncotarget
Oncotarget is a bi-weekly, peer-reviewed, open access biomedical journal covering research on all aspects of oncology.
To learn more about Oncotarget, please visit https:/
SoundCloud – https:/
Facebook – https:/
Twitter – https:/
LinkedIn – https:/
Pinterest – https:/
Reddit – https:/
Oncotarget is published by Impact Journals, LLC please visit https:/
Media Contact
[email protected]
18009220957×105
Copyright © 2021 Impact Journals, LLC
Impact Journals is a registered trademark of Impact Journals, LLC
Media Contact
Ryan James Jessup
[email protected]
Original Source
https:/
Related Journal Article
http://dx.