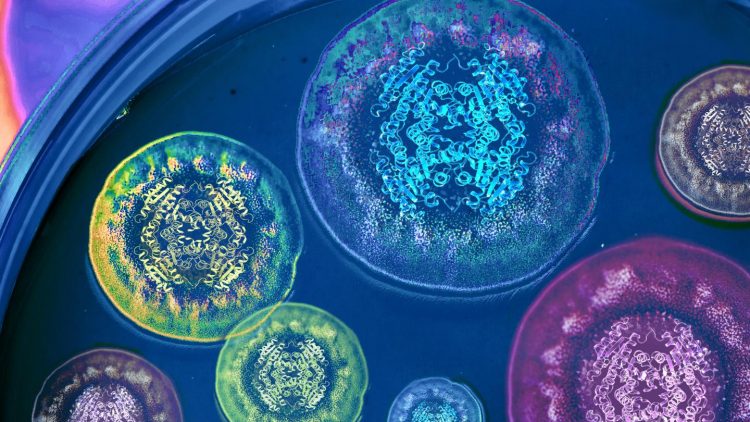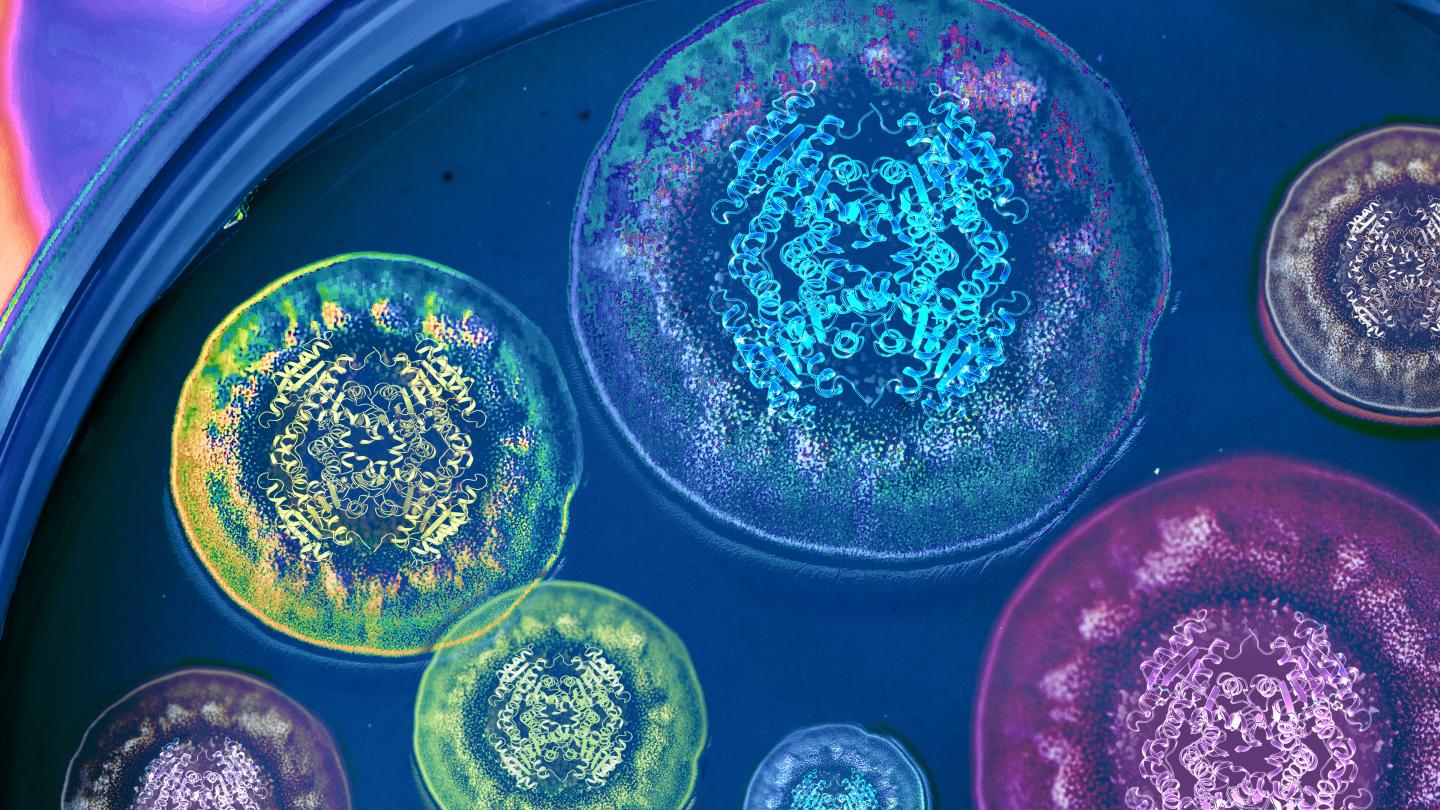
Credit: Illustration by Life Sciences Institute multimedia designer Rajani Arora.
ANN ARBOR–Research led by the University of Michigan Life Sciences Institute has solved a nearly 50-year-old mystery of how nature produces a large class of bioactive chemical compounds.
The findings are scheduled for publication on Sept. 23 in the journal Nature Chemistry.
The compounds, called prenylated indole alkaloids, were first discovered in fungi in the 1970s. Since then, they have attracted considerable interest for their wide range of potential applications as useful drugs. One compound is already used worldwide as an antiparasitic for livestock.
Understanding how the fungi build these chemicals is essential to reproducing them and creating variants in the lab for new applications. The fungi’s genes encode enzymes, and these enzymes use very simple building blocks to perform each step to build the complex molecule.
But despite the longstanding knowledge about these compounds, researchers have not been able to tease apart the precise enzymes and reactions that the fungi use to produce them.
“But if we can actually isolate the genes involved and make these enzymes, we should be able to recreate the entire bio-assembly line in a test tube,” said Qingyun Dan, a researcher in the lab of Janet Smith at the Life Sciences Institute and a lead author on the study. “But, until now, no lab has been able to do so.”
Using a collaborative approach that combined synthetic chemistry, genetics, enzymology, computational chemistry and structural biology over the course of a decade, the researchers have revealed the process–and uncovered a surprising chemical twist. The final step in the assembly process is a reaction that is nearly unprecedented in nature: the Diels-Alder reaction.
“This reaction is one of the fundamentals of synthetic organic chemistry, going back to the 1920s when it was first discovered,” said LSI faculty member David Sherman, one of the senior authors of the study.
“But even within the last few years, there has been major debate in the field about whether this reaction actually exists in nature. It’s just the most phenomenal, unexpected path to this fascinating class of indole alkaloids.”
The researchers believe that discovering the enzyme that enables the Diels-Alder reaction–and resolving how these compounds are made in nature–now opens two exciting paths forward.
First, the particular enzyme that catalyzes this Diels-Alder reaction could help improve one of the most commonly used chemical reactions. This enzyme performs the reaction with specificity that far outperforms what can commonly be achieved in the lab, meaning it creates only the one desired compound and no unintended byproducts.
Second, because the researchers were able to obtain a crystal structure of the enzyme performing the Diels-Alder reaction, they now have a clear picture of how the enzyme directs the reaction in nature–and how it might be harnessed to create new compounds in the future.
“This is a very good example of the explanatory power of crystal structures,” said Sean Newmister, a researcher in Sherman’s lab and a lead author on the study. “We gain mechanistic insight into the activity of the enzyme we’re studying, but also insight into how to use this as a tool to synthesize new chemical compounds with biological activities. And that’s really exciting.”
###
The research was supported by the National Institutes of Health and the National Science Foundation. In addition to Dan, Smith and Sherman, other study authors are: Sean Newmister, Amy Fraley, Ying Ye, Vikram Shende, Fengan Yu and W. Clay Brown of U-M; Kimberly Klas, Timothy McAfoos, Amber Somoza, James Sunderhaus, Le Zhao, Robert Paton and Robert Williams of Colorado State University; and Jacob Sanders and K.N. Houk of the University of California, Los Angeles.
The Nature Chemistry paper is titled “Fungal indole alkaloid biogenesis through evolution of a bifunctional reductase/Diels-Alderase,” DOI: 10.1038/s41557-019-0326-6.
Media Contact
Emily Kagey
[email protected]
Related Journal Article
http://dx.