Drug targets common across three coronavirus strains could be used for rapid treatment response against emerging coronavirus strains
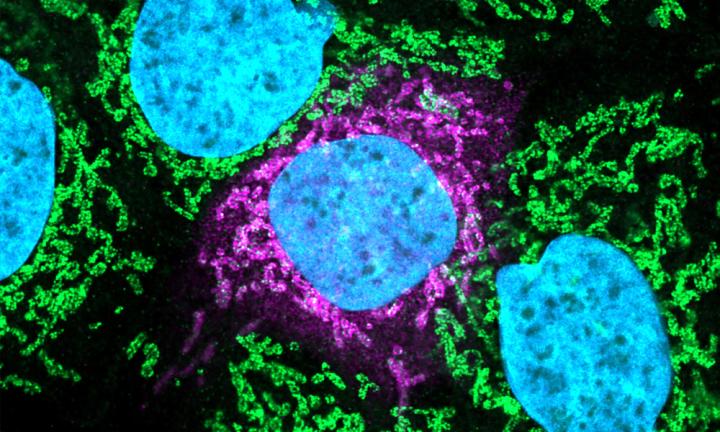
Credit: Image: Svenja Ulferts, University of Freiburg
Editing: Spencer Phillips, EMBL-EBI
15 October, Cambridge – A large international consortium of almost 200 researchers from 14 leading institutions in six countries has studied three different coronaviruses – SARS-CoV-1, SARS-CoV-2, and MERS-CoV – with the aim of finding vulnerabilities shared by these three pathogens. The research, published in the journal Science, identifies important molecular mechanisms crucial for all three coronaviruses, as well as potential drugs that could be repurposed as pan-coronavirus treatments.
The consortium included researchers at EMBL’s European Bioinformatics Institute (EMBL-EBI), the Quantitative Biosciences Institute (QBI) Coronavirus Research Group (QCRG) at University of California San Francisco (UCSF), Gladstone Institutes, Institut Pasteur, Cluster of Excellence CIBSS at the University of Freiburg, the Howard Hughes Medical Institute, and other collaborators including the biotechnology companies Aetion and Synthego.
There are three known human respiratory syndromes associated with coronaviruses: severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), and coronavirus disease 2019 (COVID-19). These are caused by SARS-CoV-1, MERS-CoV, and SARS-CoV-2, respectively.
The scientists identified drug targets and repurposed therapeutics that may have broad-spectrum activity across all three coronavirus strains. Repurposed therapeutics with known safety profiles may offer a rapid treatment response against emerging coronavirus strains in the future.
Identifying coronavirus drug targets
Building on their previous work published in Nature and Cell, the scientists determined how viral and human proteins interact, and where viral proteins are located within host cells infected by different coronaviruses. They subsequently used this data and functional genetic screening to identify host factors that prevent coronavirus propagation. The data analysed in this study will be made freely accessible through the COVID-19 Data Portal.
“These analyses demonstrate how biological and molecular information are translated into real-world implications for the treatment of COVID-19 and other viral diseases,” says Pedro Beltrao, Group Leader at EMBL-EBI. “After more than a century of relatively harmless coronaviruses, in the last 20 years we’ve had three coronaviruses that have been deadly. By looking across the species, we have the capability to predict pan-coronavirus therapeutics that may be effective in treating the current pandemic, which we believe will also offer promising therapeutics for a future coronavirus as well.”
Another step to treat COVID-19
The researchers also performed real-world analysis on clinical data regarding COVID-19 patient outcomes. To do this, they identified molecules in human cells that could be targeted with FDA-approved therapeutics and looked to see what effect these drugs had on COVID-19 patients in the clinic. This analysis involved over 740 000 patients in the United States with known SARS-CoV-2 infection.
The data and analysis carried out in this study demonstrate how molecular information can be translated into real-world implications for the treatment of COVID-19. This study also showcases a collaborative approach that can be applied to study other infectious agents in the future.
“This far-reaching international study elucidates for the first time commonalities and, importantly, vulnerabilities, across coronaviruses, including our current challenge with the COVID-19 pandemic,” says Nevan Krogan, Director of QBI and Senior Investigator at Gladstone Institutes. “In unique and rapid fashion, we were able to bridge biological and functional insights with clinical outcomes, providing an exemplary model of a differentiated way to conduct research into any disease, rapidly identify promising treatments and advance knowledge in the fields of both science and medicine. This body of work was only made possible through the collaborative efforts of senior scientific thought leaders and the teams of next-generation researchers at premier institutions across the globe.”
###
Source article
GORDON, D.E., et al. (2020). Comparative Host-Coronavirus Protein Interaction Networks Reveal Pan-Viral Disease Mechanisms. Science,. Published online 15 10; DOI: 10.1126/science.abe9403
Funding
This work was funded by grants from the National Institute of Mental Health and the National Institute of Allergy and Infectious Diseases, both part of the National Institutes of Health; the Defense Advanced Research Projects Agency; the Center for Research for Influenza Pathogenesis; the Centers of Excellence for Influenza Research and Surveillance of the National Institute of Allergy and Infectious Diseases; the Centers of Excellence for Integrative Biology of Emerging Infectious Diseases of the Agence Nationale de la Recherche (France); F. Hoffmann-LaRoche AG; Vir Biotechnology, Centre for Integrative Biological Signalling Studies (CIBSS), European Research Council (ERC) and the Ron Conway Family. A complete list of authors and full funding information is available in the Science paper.
European Bioinformatics Institute (EMBL-EBI)
The European Bioinformatics Institute (EMBL-EBI) is a global leader in the storage, analysis and dissemination of large biological datasets. We help scientists realise the potential of big data by enhancing their ability to exploit complex information to make discoveries that benefit humankind.
We are at the forefront of computational biology research, with work spanning sequence analysis methods, multi-dimensional statistical analysis and data-driven biological discovery, from plant biology to mammalian development and disease.
We are part of EMBL and are located on the Wellcome Genome Campus, one of the world’s largest concentrations of scientific and technical expertise in genomics.
Website: http://www.
Media Contact
Vicky Hatch
[email protected]
Related Journal Article
http://dx.




