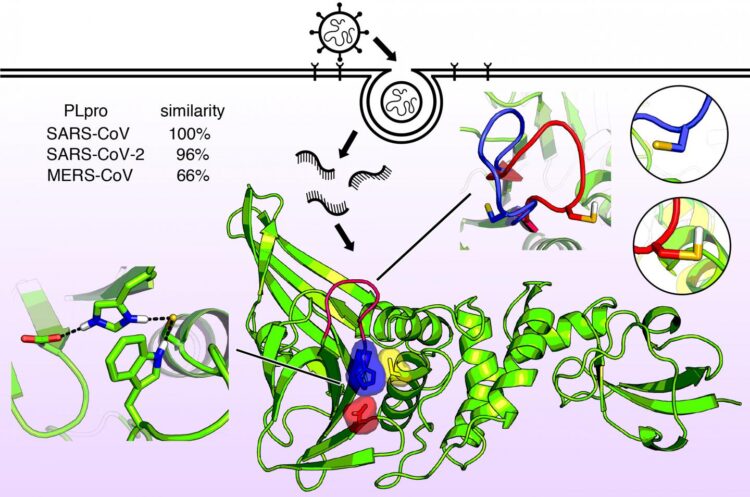
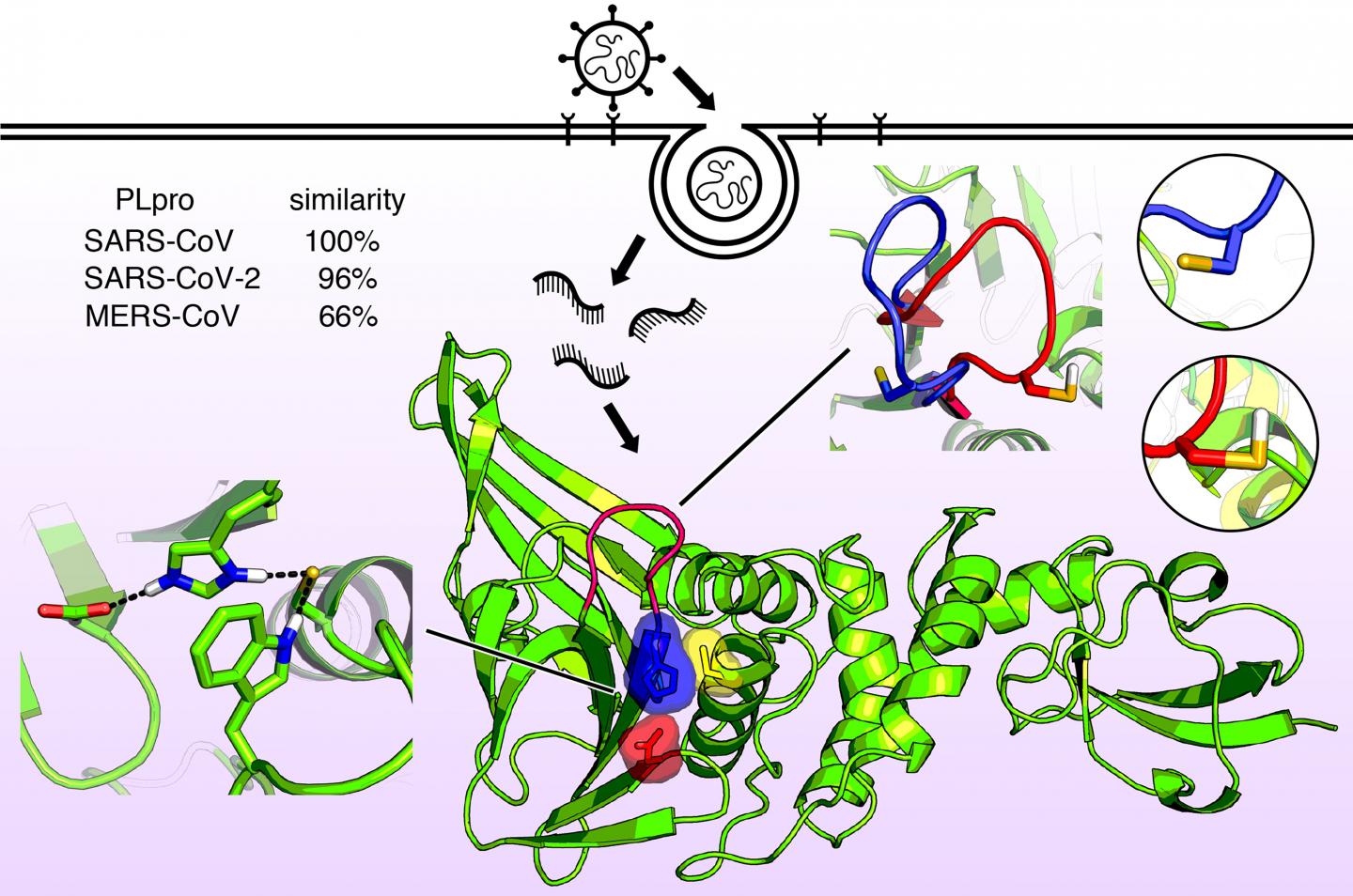
Information about the molecular shape of the viruses that cause COVID-19, SARS, and MERS reveals structural similarities that suggest possible drug treatments
Credit: Jack Henderson, University of Maryland School of Pharmacy
WASHINGTON, September 15, 2020 — COVID-19 is caused by a virus known as SARS-CoV-2, which is similar in structure to two other viruses that have caused recent outbreaks: SARS-CoV, which caused an outbreak of SARS in 2003, and MERS-CoV, the cause of a 2012 outbreak of Middle East Respiratory Syndrome.
In The Journal of Chemical Physics, by AIP Publishing, scientists from the University of Maryland School of Pharmacy report molecular-level investigations of these three viruses, providing a possible pathway to new antiviral drugs to fight all three diseases. At the present time, no effective treatment or drugs exist for any of these coronavirus diseases.
The investigators looked at a viral protein that plays a key role in the ability of the virus to replicate itself once inside the body. This protein also plays a role in defeating the host’s immune system, so it provides a particularly attractive target for potential drug treatments.
The protein, an enzyme known as the papainlike protease, PLPro, is nearly identical in SARS-CoV-2 and SARS-CoV but is slightly different in MERS-CoV. Very recently, the first structural X-ray of this enzyme revealed a shape in the catalytic domain somewhat like a hand with a “thumb,” “palm,” and “fingers.”
The thumb and palm come together to form a binding site, where a drug molecule could potentially be captured. The fingers fold down over this region and provide structural integrity that is essential for PLPro activity.
The investigators discovered small shifts in pH could change the shape of this enzyme through a process known as protonation, where hydrogen ions bind to certain amino acid units in the protein.
“Protonation state switch is an important energy transduction mechanism,” said author Jana Shen.
Co-author Jack Henderson said, “The coronavirus spike protein, for example, makes use of protonation state switches to induce large conformational changes required for membrane fusion.”
Membrane fusion is the first step in infection. A virus attaches to the outer membrane of a cell, making its way inside where it can begin to form copies of itself that spread throughout the body.
Another key feature of the PLpro binding site is a string of amino acid units called the BL2 loop. The investigators found this loop can open or close in SARS viruses when a particular amino acid on the loop is either protonated or deprotonated. In the MERS virus, however, the loop is flexible even without such an amino acid.
This feature suggests a potential drug could target the BL2 loop, causing it to close and tightly bind to a viral inhibitor.
“Our work provides a starting point for further mechanistic investigations using higher-level approaches,” said Shen.
###
The article, “Assessment of proton-coupled conformational dynamics of SARS and MERS coronavirus papain-like proteases: Implication for designing broad-spectrum antiviral inhibitors,” is authored by Jack A. Henderson, Neha Verma, Robert C. Harris, Ruibin Liu, and Jana Shen. The article will appear in The Journal of Chemical Physics on Sept. 15, 2020 (DOI: 10.1063/5.0020458). After that date, it can be accessed at https:/
ABOUT THE JOURNAL
The Journal of Chemical Physics is an international journal that publishes cutting edge research in all areas of modern physical chemistry and chemical physics. See https:/
Media Contact
Larry Frum
[email protected]
Related Journal Article
http://dx.