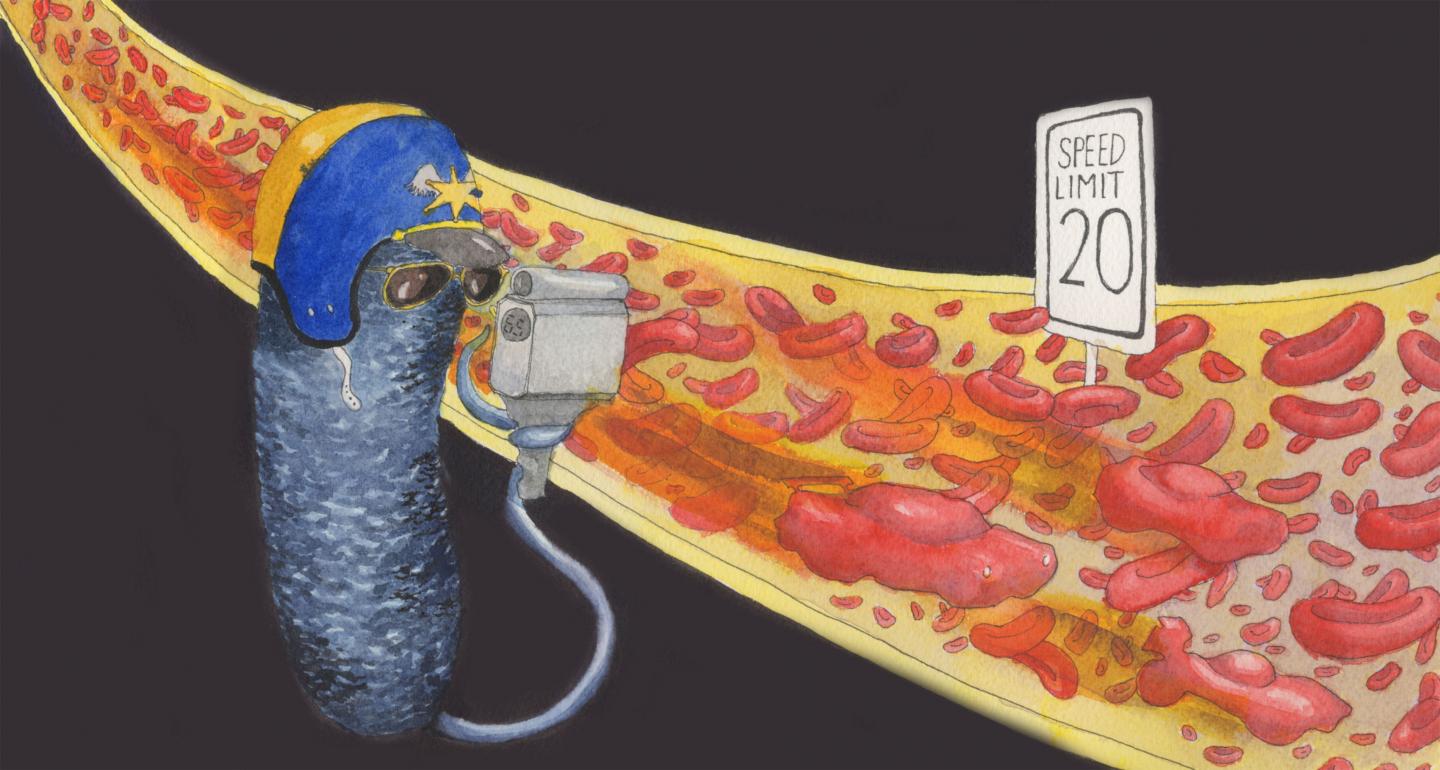
Pseudomonas bacteria detect and respond to the speed (shear rate) of flowing fluids
Credit: Illustration by Matilda Luk, Princeton University Office of Communications
An all-Princeton research team has identified bacteria that can detect the speed of flowing fluids.
Many kinds of cells can sense flow, just as our skin cells can feel the difference between a gentle breeze and a strong wind. But we depend on feeling the force involved, the push-back from the air against us. Without that push, we can’t distinguish speed; when the windows are closed, our skin can’t feel any difference in air force whether we are sitting in an office, a speeding car or a cruising airplane. But now, a team of Princeton researchers has now discovered that some bacteria can in fact detect the speed of flow regardless of the force. Their paper appears in the online journal Nature Microbiology.
“We have engineered bacteria to be speedometers,” said Zemer Gitai, Princeton’s Edwin Grant Conklin Professor of Biology and the senior author on the paper. “There’s an application here: We can actually use these bacteria as flow sensors. If you wanted to know the speed of something in real time, we can tell you.”
The bacterium with the built-in speedometer, Pseudomonas aeruginosa, is a ubiquitous pathogen, found in and on bodies, in streams of water, in soil, and throughout hospitals. The Center for Disease Control and Prevention classifies Pseudomonas as a “serious threat,” responsible for more than 50,000 healthcare-associated infections per year, of which 6,700 are antibiotic resistant and 440 are fatal.
“In some hospitals, surgeons use chemical disinfectants — Purell, essentially — instead of scrubbing in because Pseudomonas loves to grow in pipes,” said Gitai. In fact, Pseudomonas is nearly always surrounded by flowing fluids, whether in a stream or in any of the human body systems that involve flow — the blood stream, the urinary tract, the gastrointestinal tract, even the lungs — or in plumbing systems or medical equipment like catheters, one of the primary vectors for post-surgical infections.
“That’s old news,” said Gitai. “What we found is that not only do Pseudomonas encounter flow, but that they actually can sense and respond to that flow. That’s a big deal. If they’re in flow, they can change their ‘behavior,’ if you will, based on feeling they’re in flow.”
Joseph Sanfilippo, a postdoctoral research associate in Gitai’s lab, and Alexander Lorestani, a 2017 graduate alumnus, are the lead authors on the paper. Together, they determined that bacteria can detect and even measure the flow speed of nearby fluids to turn on a set of genes. They called those genes “fro,” for “flow-regulated operon.”
“Fro’s response is not just an on-off switch; it’s actually tuned to the speed,” said Sanfilippo. “It’s more like a dimmer switch than a light switch.”
They then bioengineered a connection between fro and a gene that causes Pseudomonas to glow, so that fro’s genetic response could be seen under a microscope, thereby creating a real-time visual speedometer. The faster the flow, the brighter the glow.
Using that tool, Sanfilippo was able to determine the range of speeds that Pseudomonas responds to, with a surprising result: “They turn out to perfectly coincide with the range of speeds of fluids that are known in the blood stream and the urinary tract,” said Gitai.
Given the microscopic scale of the bacteria and the strand-of-hair-sized flow chambers in which they tested fro (50 microns tall by 500 microns wide), the researchers did not measure speed in traditional miles per hour (or meters per second) units, but instead measured “shear rate,” the rate at which adjacent layers of fluid pass one another. That is measured in distance-less “per-second” units.
They found that fro did not respond to shear rates below 8 per-second — slower than most fluids found in the human body — but that it did tune its response to shear rates between 40 and 400 per-second, then plateaued above that. For reference, the shear rate in average-sized human veins is about 100 per-second.
“The speeds that fro responds to are the speeds that are going through your body right now,” said Sanfilippo.
“The authors combine their expertise in bioengineering and biophysics (to fabricate these nifty flow chambers) with cutting-edge biology (RNA sequencing) to understand some fundamental questions in microbiology,” said Joanne Engel, chief of infectious disease and a professor of microbiology and immunology at the University of California-San Francisco, who was not involved in this research.
“The ability to bring investigators together from diverse fields helps to push research forward in new and creative ways,” she said. “As this sensing mechanism is likely to be found throughout biology, it may have broad applications and may even be useful for developing new drugs (antibiotics) to treat bacterial infections, especially blood infections (like sepsis).”
A force-independent response
Sanfilippo and his colleagues had initially assumed that Pseudomonas’s sensitivity to flow must depend on its ability to sense force, just as we all intuitively understand that a hand out a car window will feel more force when we’re driving at higher speeds. That intuitive link between flow and force has led to a universal blind spot in the field, said Gitai.
“Other researchers have found that different bacteria can respond to fluid flow, and they’ve effectively assumed that it was the force,” Gitai said. “The intuition was so strong that it should be force, that in fact people didn’t bother to explicitly test this.”
A way to test that assumption came from Howard Stone, the Donald R. Dixon ’69 and Elizabeth W. Dixon Professor of Mechanical and Aerospace Engineering and a co-author on the paper. Stone proposed an experiment that would show whether or not the bacteria was responding to force: submit it to materials of different viscosities, or thicknesses, that are flowing at the same speed.
If you pour water over your hand, then pour honey over it at the same speed, the more viscous honey will push harder against your skin, so your hand will feel more force. Sanfilippo ran experiments with fluids up to 10 times more viscous than the default medium and found that fro responded only to the shear rate — the speed — not the force of the thicker fluids.
“Doing that one additional experiment — we call it Howard’s experiment — really was key,” said Sanfilippo. That enabled the researchers to say conclusively that Pseudomonas was tuning its responses purely to the speed of the flowing materials, something that has never before been documented. To distinguish this from other kinds of mechanical sensing that have been documented in bacteria, the team coined the term “rheosensing,” or flow-sensing, to describe the phenomenon.
“We are launching a mini-field here,” said Gitai. “Thinking about how bacteria live in flow is a completely under-explored area. We’d love for people to look at this with other bacteria. And as we said, there’s been a huge assumption in mammalian studies that everything is force-dependent — we’d love for people to read our paper and then go back and revisit some of those assumptions and change the viscosity in their systems.”
It’s still not clear how Pseudomonas benefits from knowing the speed of its surrounding fluids, said Gitai, but it’s probably linked to the pathogen’s versatility; he calls it the Swiss Army knife of bacteria.
“Pseudomonas has a huge arsenal of different ways that it can attack,” Gitai said. “One idea is that it wants to know when it’s appropriate to use which tool in its arsenal. If you have a burn, and it’s just sitting on your skin, maybe the genes that it can use to hurt you are different than the genes that it would use to hurt you in your urinary tract or in your blood. And that’s the big idea here, that — to anthropomorphize it — it can know if there’s flow around, and that may help it tune its response to its environment.”
The all-Princeton research team also included Matthias Koch, a postdoctoral research associate in the Lewis-Sigler Institute for Integrative Genomics; Benjamin Bratton, an associate research scholar in molecular biology; and Albert Siryaporn, a former postdoctoral researcher in Gitai’s lab who is now an assistant professor at the University of California-Irvine.
###
“Microfluidic-based transcriptomics reveal force-independent bacterial rheosensing,” by Joseph E. Sanfilippo, Alexander Lorestani, Matthias D. Koch, Benjamin P. Bratton, Albert Siryaporn, Howard A. Stone and Zemer Gitai, was released online on May 13 by Nature Microbiology (DOI: 10.1038/s41564-019-0455-0). This work was supported by the National Institutes of Health (grants DP1AI124669, K22AI112816 and R21AI121828), the National Science Foundation (PHY-1734030), the Glenn Centers for Aging Research and the German Research Foundation (DFG Award KO5239/1-1).
Media Contact
Liz Fuller-Wright
[email protected]
Related Journal Article
http://dx.




