Induced pluripotent stem cells hold promise for regenerative medicine because they can, in theory, turn into any type of tissue and because they are made from a patient’s own adult cells, guaranteeing compatibility. However, the technique that turns adult cells into these iPS cells is not foolproof; after reverting to their pluripotent state, these cells don’t always correctly differentiate back into adult cells.
Researchers from the University of Pennsylvania have now discovered one of the reasons why: the reversion process does not always fully capture the way a cell’s genome is folded up inside its nucleus. This folding configuration directly influences gene expression and therefore the functionality of the cell.
The new study shows that current techniques might not produce iPS cells that are equivalent to the pluripotent stem cells found in embryos, as some clones retain folding patterns that partially resemble those found in the adult cells from which they are made.
Led by Jennifer Phillips-Cremins, assistant professor in the School of Engineering and Applied Science’s Department of Bioengineering, and Jonathan Beagan, a graduate student in her lab, the study, published in the journal Cell Stem Cell, also suggests ways of minimizing these folding errors.
Though techniques for reverting adult cells into iPS cells have existed for a decade and avoid the issues surrounding the use of embryonic stem cells that have stymied research into regenerative medicine, clinical investigations of these cells have been cautious and slow. IPS cells can fail to correctly differentiate into the desired tissue. Moreover, there are also concerns that the resulting tissue could have unforeseen genetic abnormalities or could become cancerous.
Even outside the clinical applications, many researchers are interested in iPS cells as a way of generating a “disease in a dish.” Rather than taking a tissue sample from a patient with a genetic disorder, which is especially challenging when the affected organ is the brain, researchers could use iPS cells derived from that patient’s skin cells to grow model organs as needed. Observing the development of those tissues could provide clues to the progression of the disease, as well as serve as ideal test-beds for treatments not yet approved for use in humans.
In both clinical and research applications, however, the traits that allow for the generation of “high quality” iPS cells capable of correctly differentiating into the desired tissue with no genetic abnormalities is unclear.
“We know there is a link between the topology of the genome and gene expression,” Phillips-Cremins said, “so this motivated us to explore how the genetic material is reconfigured in three dimensions inside the nucleus during the reprogramming of mature brain cells to pluripotency. We found evidence for sophisticated configurations that differ in important ways between iPS cells and embryonic stem cells.”
Phillips-Cremins’ area of research is “3-D epigenetics,” or the way that the folding of DNA influences gene expression. Classic epigenetic marks are chemical modifications on top of the DNA sequence that provide an additional layer of information on top of the long sequence of base-pair letters. Looking at these marks in a linear fashion does not reveal the whole picture, however, as genome folding can bring two disparate regions of the DNA into spatial and functional contact.
By applying experimental and computational techniques that the Cremins lab has developed, her group was able to identify folding patterns in iPS cells that had previously been unseen.
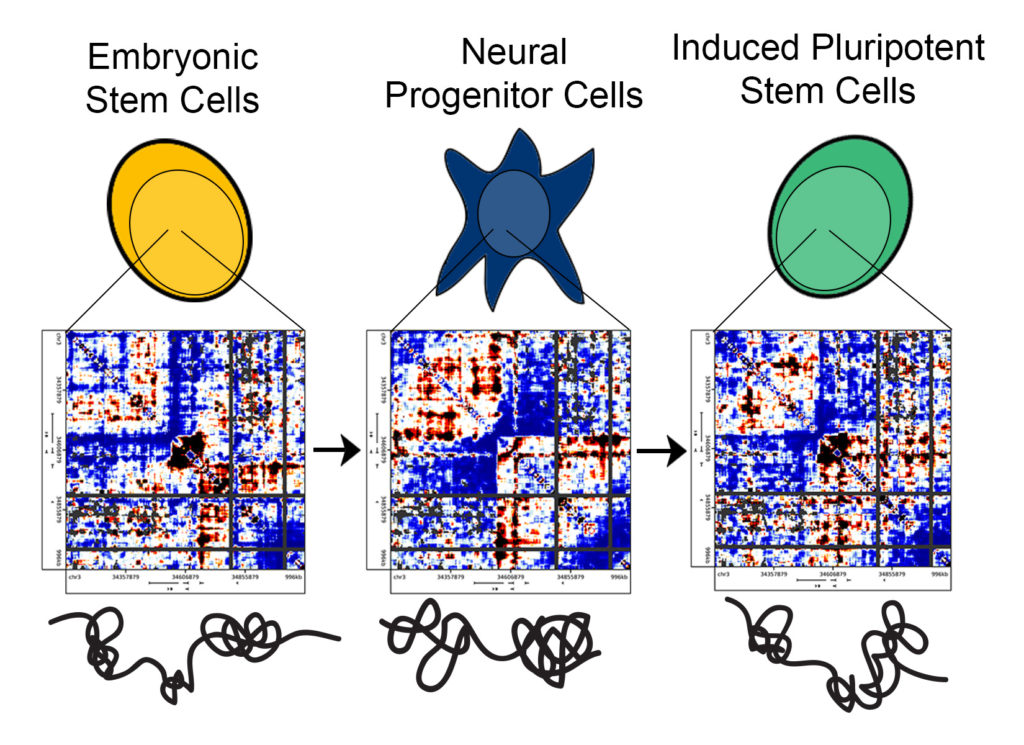
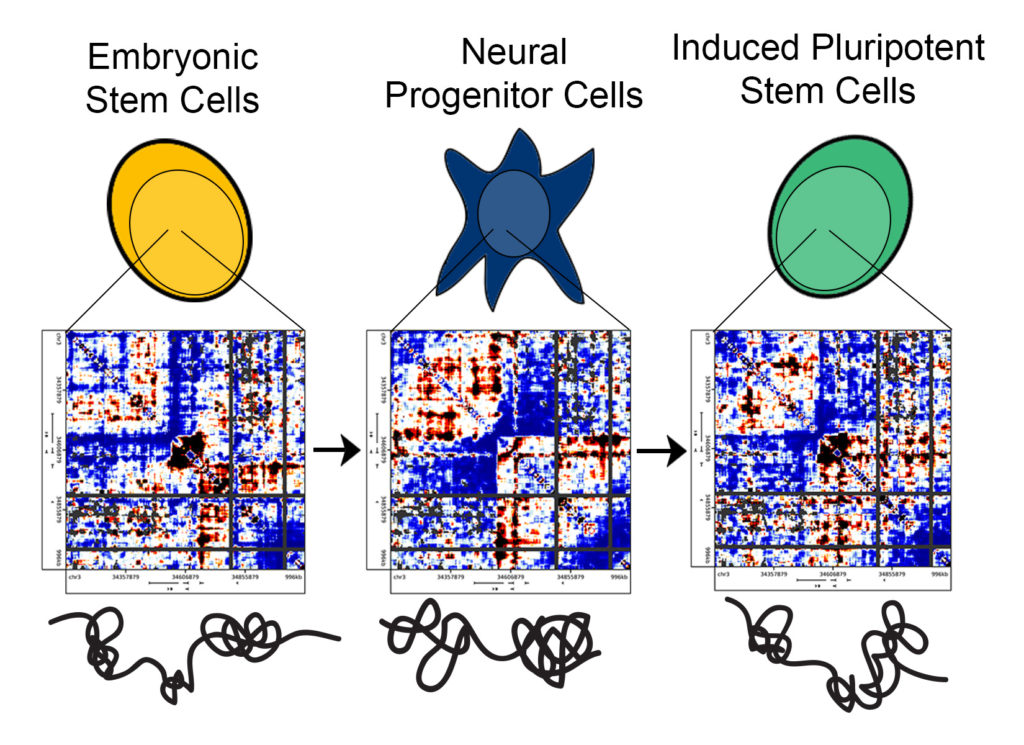
“Previous methods provided data analogous to an old television, with large, black-and-white pixels,” Phillips-Cremins said. “One could generate a blurry image and tell there was a person on the screen, but it would be difficult to parse finer-scale facial features. We employed methods to create high-resolution maps of genome folding, so we could distinguish detailed topological features and evaluate their similarities and differences among traditional embryonic stem cells, iPS cells and mature, differentiated cells.”
The approach Beagan used to create the high-resolution maps involves fixing the DNA such that its 3-D folding patterns are preserved prior to sequencing. Sections of the linear genetic sequence that are separated by marked distances but are spatially adjacent when the DNA is folded are chemically glued together. As a result, two distant parts of the linear sequence will end up in the same string of hybrid DNA and will thus be detected together when the DNA is sequenced.
Analyzing these hybrid pieces provides information that allows the researchers to infer which DNA segments are adjacent to each other in the genome’s folded state. Critically, the Cremins lab’s approach targets only specific sites in the genome, which allows high-resolution analysis across these regions to be much more easily achievable than with alternative genome-wide approaches. The maps reported by the Penn Engineering lab are the highest-resolution maps of genome folding to date in iPS cells.
Phillips-Cremins and her computational team can plot the sequencing data in heat maps, thereby providing a picture of the DNA segments that are spatially adjacent to each other in the 3-D nucleus of stem cells.
“Each of the pixels on our heat maps,” Beagan said, “is a representation of the frequency that any two given segments in the genome interact. You can represent the contact frequency numbers as colors and the entire region of DNA as a heat map. You end up seeing intriguing patterns of high and low intensity, and from these patterns one can infer the folding configurations of the genome.”
The Penn researchers targeted several locations along the genome to perform their computational analyses, comparing the iPS cells to the cells from which they were generated and to the embryonic stem cells they ideally should perfectly replicate.
They found that traditional embryonic stem cells and mature, differentiated brain cells had strikingly different genome folding patterns. Surprisingly, however, the genetic material from iPS cells did not fold in a manner that perfectly resembled traditional embryonic stem cells but instead exhibited traces of the 3-D configurations of the brain cells from which they were derived.
“We found marked differences among the heatmaps we generated for each cell type,” Beagan said. “Our observations are important because they suggest that, if we can push the 3-D genome conformation of cells that we are turning into iPS cells to be closer to that of embryonic stem cells, then we can possibly generate iPS cells that match gold-standard pluripotent stem cells more rapidly and efficiently.”
Importantly, the researchers also found they could make iPS cells with more accurate folding configurations by changing the medium in which they are grown. This result provides a window of insight into possible ways that researchers and clinicians could better engineer iPS cells for experiments and treatments. The Phillips-Cremins lab already has experiments underway to engineer 3-D genome folding in stem cells to control gene expression on demand.
###
The research was supported by The New York Stem Cell Foundation, the Alfred P. Sloan Foundation, the NIH Director’s New Innovator Award from the National Institute of Mental Health (1DP2MH11024701) and a 4D Nucleome Common Fund grant (1U01HL12999801).
Other researchers contributing to the study include Phillips-Cremins lab members Thomas G. Gilgenast, Jesi Kim, Zachary Plona, Heidi Norton and Gui Hu; Sarah Hsu and Emily Shields of Penn’s Perelman School of Medicine epigenetics program; biologists Xiaowen Lyu and Victor G. Corces of Emory University; Effie Apostolou and Konrad Hochedlinger of the Massachusetts General Hospital Cancer Center and Center for Regenerative Medicine; and Job Dekker, a Howard Hughes Medical Institute researcher. Apostolou is now affiliated with the Meyer Cancer Center and Department of Medicine at Weill Cornell Medical College.
Media Contact
Evan Lerner
[email protected]
215-573-6604
@Penn
http://www.upenn.edu/pennnews
The post Penn bioengineers show why lab-made stem cells might fail: Errors in DNA folding appeared first on Scienmag.