Rice scientists analyze structure, mechanism of phage protein that steals electrons
Credit: Illustration by Ian Campbell/Rice University
HOUSTON – (May 22, 2020) – Beneath the ocean’s surface, a virus is hijacking the metabolism of the most abundant organism on Earth. That may be of interest to those of us above who breathe.
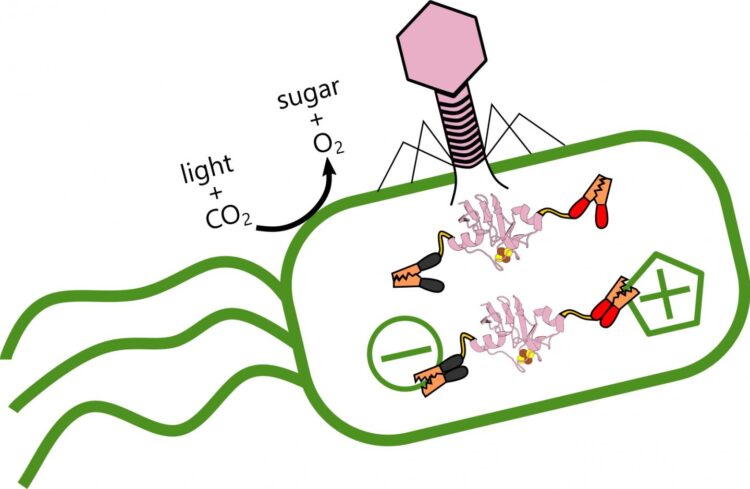
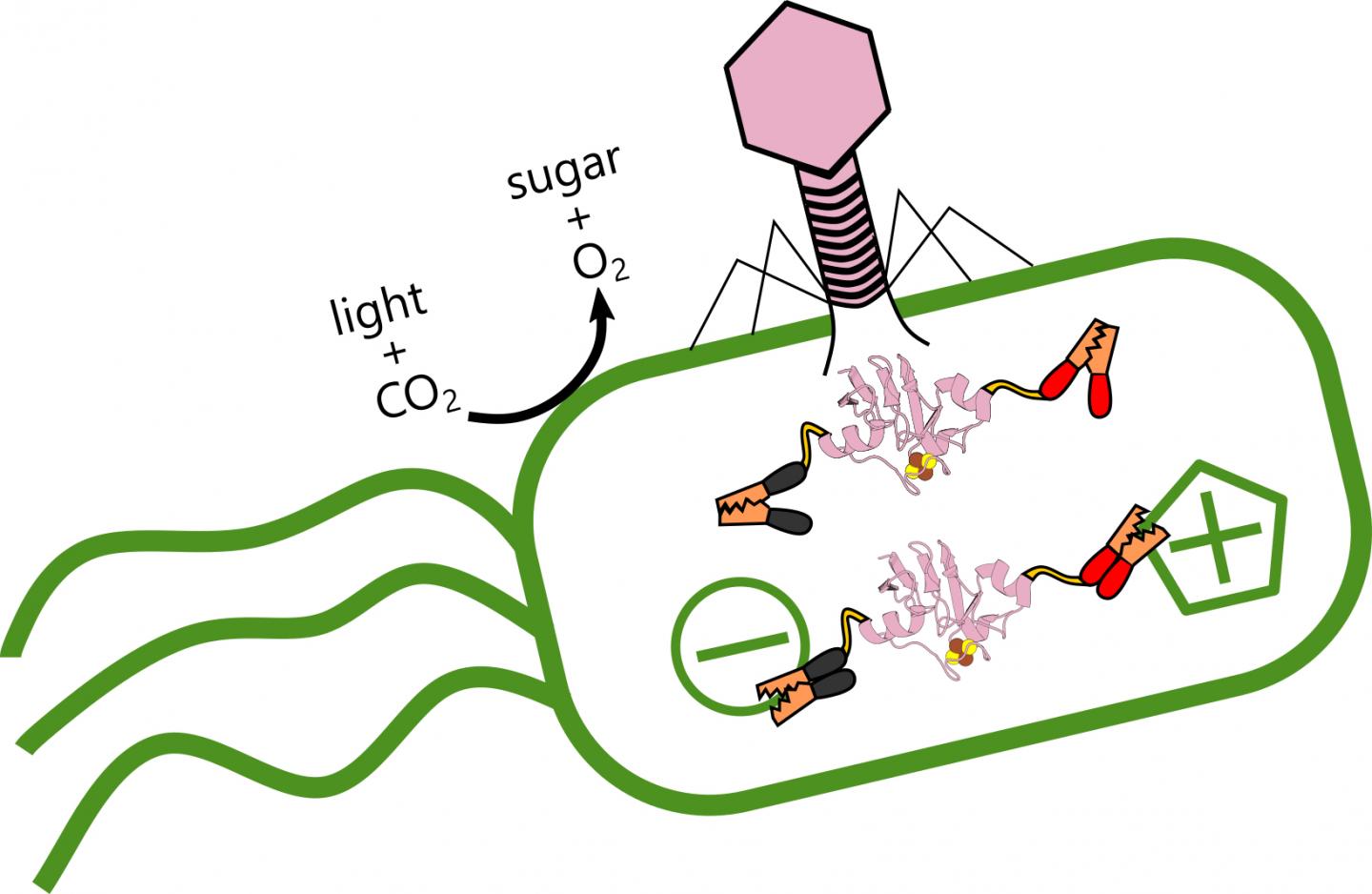
Rice University scientists analyzed the role of ferredoxin proteins produced when phages alter the ability of Prochlorococcus marinus to store carbon and counter the greenhouse gas effect arising from fossil fuel consumption.
P. marinus is a photosynthetic cyanobacteria that resides primarily in the tropics and subtropics, where an estimated 10-to-the-27 (an octillion) of them use sunlight to produce oxygen and collectively store four gigatons of carbon annually. Some of this carbon provides critical feedstocks for other marine organisms.
But phages are not their friends. The virus strengthens itself by stealing energy the bacteria produces from light, reprogramming its victim’s genome to alter how it transfers electrons.
P. marinus and its carbon-storing mechanism are sensitive to temperature, so it bears watching as climate change warms the oceans and extends its range, said Ian Campbell, a Rice postdoctoral researcher and lead author of the study in the Journal of Biological Chemistry.
“The growth in the range of this organism in the oceans could increase the total carbon stored by these microbes,” he said. “Alternatively, the viruses that infect these bacteria could alter carbon fixation and potentially prevent gigatons of carbon from being taken out of the air annually, according to one recent projection.”
Campbell said the goal of the study was to explore the variety of ways viruses interact with their hosts. In the process, the researchers discovered the phage wrests control of electron flow in the host itself, rewiring the bacteria’s metabolism. “When the virus infects, it shuts down production of the bacterial proteins and replaces it with its own variants,” he said. “I compare it to putting a different operating system in a computer.”
The researchers used synthetic biology techniques to mix and match phage and cyanobacterial proteins to study how they interact. A part of the study led by Rice biochemist George Phillips also determined for the first time the structure of a key cyanophage ferredoxin protein.
“A phage would usually go into a cell and kill everything,” said Rice synthetic biologist Jonathan Silberg, the study’s lead scientist and director of the university’s Systems, Synthetic and Physical Biology program.
“But Ian’s results suggest these phages are establishing a complex control mechanism,” he said. “I wouldn’t say they’ve zombified their hosts, because they allow the cells to continue doing some of their own housekeeping. But they’re also plugging in their own ferredoxins, like power cables, to fine tune the electron flow.”
Instead of working directly with cyanophages and P. marinus, Campbell and his team used synthetic biology tools to reprogram much larger, better-understood Escherichia coli bacteria to express genes that mimicked interactions between the two.
“Taking a phage and a cyanobacteria from the ocean and trying to study the biology, especially electron flow, would be really hard to do through classical biochemistry,” Silberg said. “Ian literally took partners from both the phage and the host, put them together by encoding their DNA in another cellular system, and was able to quickly develop some interesting results.
“It’s an interesting application of synthetic biology to understand complex things that would otherwise be arduous to measure,” he said.
The researchers suspect the protein they modeled in E. coli, the Prochlorococcus P-SSM2 phage ferredoxin, is nothing new. “People knew phages encode different things that do electron transfer, but they didn’t know how to connect the wires between the phage and the host,” Silberg said. “They also didn’t know a lot about the phage’s evolution. The structure makes it clear this phage can be traced to specific ancestral proteins involved in photosynthesis.”
###
Co-authors of the paper are graduate student Jose Luis Olmos Jr., research technician Weijun Xu, postdoctoral researchers Dimithree Kahanda and Joshua Atkinson, undergraduate alumnus Othneil Noble Sparks, research scientist Mitchell Miller, and George Bennett, the E. Dell Butcher Professor of BioSciences and a professor of chemical and biomolecular engineering. Phillips is a professor of biosciences. Silberg is the Stewart Memorial Professor of Biochemistry and a professor of biosciences, bioengineering, and chemical and biomolecular engineering.
The Department of Energy, NASA, the National Science Foundation, the Moore Foundation, the National Cancer Institute and the National Institute of General Medical Sciences supported the research.
Read the abstract at https:/
This news release can be found online at https:/
Follow Rice News and Media Relations via Twitter @RiceUNews.
Related materials:
Labs give ancient proteins new purpose: http://news.
Silberg Lab: https:/
Rice Department of Biosciences: https:/
Wiess School of Natural Sciences: https:/
Systems, Synthetic and Physical Biology graduate program: https:/
Images for download:
https:/
Rice University scientists are analyzing the role of ferredoxin proteins produced when viral phages alter electron transfer in ocean-dwelling, photosynthetic bacteria that produce oxygen and store carbon. When the virus (pink) infects the bacteria, it produces a ferredoxin protein that hooks into the bacteria’s existing electrical structure and alters its metabolism. (Credit: Illustration by Ian Campbell/Rice University)
https:/
Rice University synthetic biologist Jonathan Silberg, left, and postdoctoral researcher Ian Campbell led a team that analyzed the role of ferredoxin proteins produced when viral phages alter electron transfer in ocean-dwelling, photosynthetic bacteria that produce oxygen and store carbon. (Credit: Jeff Fitlow/Rice University)
Located on a 300-acre forested campus in Houston, Rice University is consistently ranked among the nation’s top 20 universities by U.S. News & World Report. Rice has highly respected schools of Architecture, Business, Continuing Studies, Engineering, Humanities, Music, Natural Sciences and Social Sciences and is home to the Baker Institute for Public Policy. With 3,962 undergraduates and 3,027 graduate students, Rice’s undergraduate student-to-faculty ratio is just under 6-to-1. Its residential college system builds close-knit communities and lifelong friendships, just one reason why Rice is ranked No. 1 for lots of race/class interaction and No. 4 for quality of life by the Princeton Review. Rice is also rated as a best value among private universities by Kiplinger’s Personal Finance.
Contacts:
Jeff Falk
713-348-6775
[email protected]
Jade Boyd
713-348-6778
[email protected]
Media Contact
Jade Boyd
[email protected]
Original Source
https:/
Related Journal Article
http://dx.