Credit: Image courtesy of Peter Hoey.
Bacteria are everywhere. They live in the soil and water, on our skin and in our bodies. Some are pathogenic, meaning they cause disease or infection. To design effective treatments against pathogens, researchers need to know which specific genes are to blame for pathogenicity.
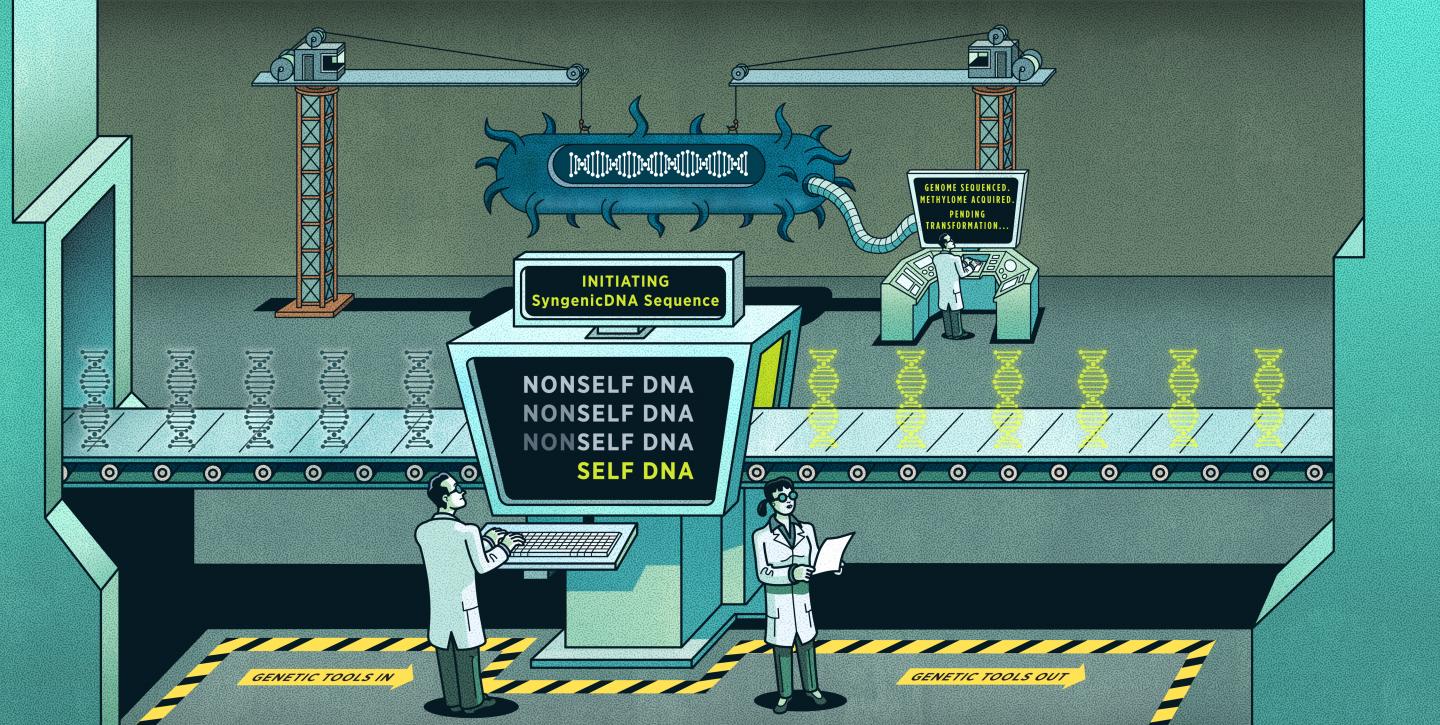
Scientists can identify pathogenic genes through genetic engineering. This involves adding human-made DNA into a bacterial cell. However, the problem is that bacteria have evolved complex defense systems to protect against foreign intruders–especially foreign DNA. Current genetic engineering approaches often disguise the human-made DNA as bacterial DNA to thwart these defenses, but the process requires highly specific modifications and is expensive and time-consuming.
In a paper published recently in the Proceedings of the National Academy of Sciences journal, Dr. Christopher Johnston and his colleagues at the Forsyth Institute describe a new technique to genetically engineer bacteria by making human-made DNA invisible to a bacterium’s defenses. In theory, the method can be applied to almost any type of bacteria.
Johnston is a researcher in the Vaccine and Infectious Disease Division at the Fred Hutchinson Cancer Research Center and lead author of the paper. He said that when a bacterial cell detects it has been penetrated by foreign DNA, it quickly destroys the trespasser. Bacteria live under constant threat of attack by a virus, so they have developed incredibly effective defenses against those threats.
The problem, Johnston explained, is that when scientists want to place human-made DNA into bacteria, they confront the exact same defense systems that protect bacteria against a virus.
To get past this barrier, scientists add specific modifications to disguise the human-made DNA and trick the bacterium into thinking the intruder is a part of its own DNA. This approach sometimes works but can take considerable time and resources.
Johnston’s strategy is different. Instead of adding a disguise to the human-made DNA, he removes a specific component of its genetic sequence called a motif. The bacterial defense system needs this motif to be present to recognize foreign DNA and mount an effective counter-attack. By removing the motif, the human-made DNA becomes essentially invisible to the bacterium’s defense system.
“Imagine a bacterium like an enemy submarine in a dry-dock, and a human-made genetic tool as your soldier that needs to get inside the submarine to carry out a specific task. The current approaches would be like disguising the spy as an enemy soldier, having them walk up to each gate, allowing the guards to check their credentials, and if all goes well, they’re in,” Johnston said. “Our approach is to make that soldier invisible and have them sneak straight through the gates, evading the guards entirely.”
This new method requires less time and fewer resources than current techniques. In the study, Johnston used Staphylococcus aureus bacteria as a model, but the underlying strategy he developed can be used to sneak past these major defense systems that exist in 80 to 90 percent of bacteria that are known today.
This new genetic engineering tool opens up the possibilities for research on bacteria that haven’t been well studied before. Since scientists have a limited amount of time and resources, they tend to work with bacteria that have already been broken into, Johnston explained. With this new tool, a major barrier to breaking into bacteria DNA has been solved, and researchers can use the method to engineer more clinically relevant bacteria.
“Bacteria are the drivers of our planet,” said Dr. Gary Borisy, a Senior Investigator at the Forsyth Institute and co-author of the paper. “The capacity to engineer bacteria has profound implications for medicine, for agriculture, for the chemical industry, and for the environment.”
###
This research was facilitated by an ‘NIH Director’s Transformative Research Award’ (R01OD024734) granted to the research team in 2017 through the National Institute of Dental and Craniofacial Research (NIDCR).
Media Contact
Alexandra Nicodemo
[email protected]
Original Source
https:/




