Credit: Aretha Fiebig
Many people have never heard of Brucellosis, but farmers and ranchers in the United States forced to cull animals that test positive for the disease and people infected by the animal-transmitted Brucella abortus (B. abortus) pathogen that suffer chronic, Malaria-type symptoms, certainly have.
Brucellosis is an agricultural and human health concern on a global scale. It was introduced over 100 years ago to Bison and elk in Yellowstone National Park by cattle and has been circulating among the wild herds ever since, leading to periodic outbreaks and reinfection. There is no vaccine for humans, and experimental studies of B. abortus in its natural animal hosts are technically difficult, extremely expensive and only a few facilities are capable of conducting these studies.
That did not stop Sean Crosson, MSU Rudolph Hugh Endowed Professor, and colleague Aretha Fiebig, research associate professor in Michigan State University’s Department of Microbiology and Molecular Genetics, from bringing sophisticated genomics tools from the lab to the field to gain new insight into how B. abortus infects cattle and help stop the spread of this deadly disease.
The results of their study were published in the Proceedings of the National Academy of Sciences.
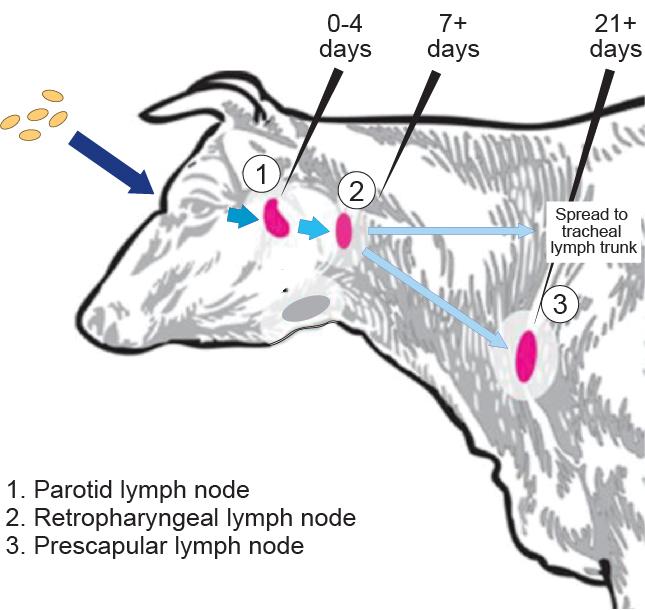
“B. abortus primarily infects cattle, causing pregnant cows to abort the fetus, but the infection is typically studied in mouse models, which are not the true host of the bacterium,” explained Crosson, who has been studying the bacteria for more than 14 years. “If you want to understand the infection biology underlying bovine disease, then it’s helpful to study things in the natural host in a field context.”
In the microbiological equivalent to tagging cattle, Crosson and Fiebig harnessed the hopping ability of specialized DNA called transposons to tag individual strains of B. abortus with unique barcodes. This gave them the ability to count how many B. abortus bacteria made it from the cow’s eye, a common point of infection in the field, to the lymph nodes.
“As molecular biologists, we can harness hopping DNA by separating away the enzyme that allows it to continue moving,” said Fiebig, who specializes in bacterial regulatory mechanisms. “We temporarily gave DNA the ability to hop into the B. abortus genome, but it didn’t hop out again.”
The scientists mixed millions of E. coli bacteria carrying transposons with millions of B. abortus in a broth containing amino acids and sugars, initiating mass tagging through a process called bacterial conjugation where transposons hop into and unite with the B. abortus genome. When the remaining E. coli cells were washed away, they were left with a vat of individually barcoded B. abortus strains.
“We were able to make a rich pool of about a million different barcoded strains,” Fiebig said. “When we infected the cattle, we could track almost every single strain and ask, ‘how many strains got lost, what strains had an advantage and was that advantage for a genetic reason or just chance?'”
Millions of bacteria went in, but surprisingly few came out. And while the genetic identity of strains that made it through was random, the number of strains that managed to infect individual lymph nodes was remarkably similar.
“We knew there was some restriction, or infection bottleneck, but we didn’t understand the magnitude until this study,” said Fiebig.
The unexpected results required nontraditional computational analysis, so Marianne Huebner, director of MSU’s Center for Statistical Training and Consulting, provided expert guidance on the use of mathematical models to assess the population structures of the bacteria that survived the bottleneck.
Infecting a large animal host with a federally regulated pathogen also presented significant methodological challenges. The researchers relied on the highly skilled veterinarians and high-tech facilities at the United States Department of Agriculture-Agricultural Research Service’s (USDA-ARS) National Animal Disease Center in Ames, Iowa where the cows in the study were housed and treated.
The challenging field study paid off, providing critical insight into just how good the cow’s mucosal barrier is at restricting B. abortus during infection.
“In the end, we gained a quantitative understanding of an infection bottleneck via a common route of bovine infection in the field,” Crosson said. “This information is useful for scientists studying the epidemiology of Brucellosis in livestock and wildlife and can help us build better transmission models as we work to stop the spread of this disease.”
The proof of principle paper also opened new doors to discovering specific B. abortus genes involved in the devastating disease.
“Going forward, we better understand how to use our library of barcoded mutants over longer timepoints in a pregnant host to find B. abortus genes that influence the most severe outcomes in cattle, including abortion,” Crosson said. “This is a goal of the USDA-ARS as well, to know what genes in B. abortus are critical to infection in the bovine host.”
###
The researchers emphasized that MSU’s College of Veterinary Medicine, the College of Natural Science and AgBioResearch in the College of Agriculture and Natural Resources were instrumental in supporting the study.
Media Contact
Caroline Brooks
@MSUnews
517-420-8376
Original Source
https:/
Related Journal Article
http://dx.




