Researchers investigated observer performance for 2D and 3D image localization tasks; they found that when observers scrolled through a 3D image, there was little evidence of integrating information in the scrolling direction
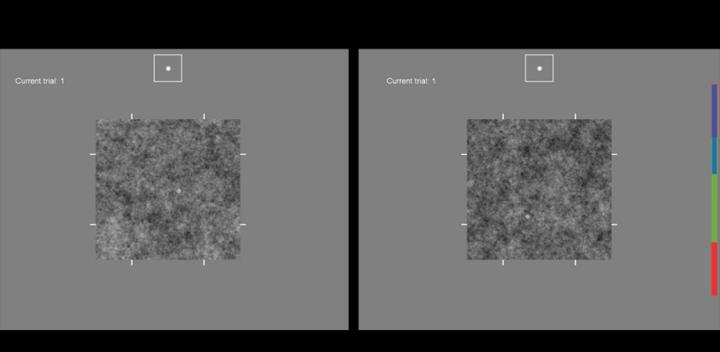
Credit: Abbey, Lago, and Eckstein (Fig. 2), doi 10.1117/1.JMI.8.4.041206
Three-dimensional or “volumetric” images are widely used in medical imaging. These images faithfully represent the 3D spatial relationships present in the body. Yet 3D images are typically displayed on a two-dimensional monitor, which creates a dimensionality mismatch that must be resolved in a clinical setting where practitioners must search a 2D or a 3D image to find a particular trait or target of interest.
To learn more about this problem, Craig K. Abbey, Miguel A. Lago, and Miguel P. Eckstein, of the Department of Psychological and Brain Sciences at University of California Santa Barbara, used techniques from the field of vision science to examine how the observers use information in images to perform a given task. Their research, published in the Journal of Medical Imaging, evaluates human performance in localization tasks which involve searching a 2D or a 3D image to find a target that is masked by noise. The addition of noise makes the task difficult, similar to reading images in a clinical setting.
The images in the study were simulations intended to approximate high-resolution x-ray computed tomography (CT) imaging. The images were generated in 3D and viewed as 2D “slices,” and the test subjects in the experiment were able to freely inspect the images, including scrolling through 3D images. Abbey explains, “Many techniques for image display have been developed
but it is not uncommon for volumetric images to be read in a clinical setting by simply scrolling through a ‘stack’ of 2D sections.” The study aimed to compare components of reader performance in 3D images to 2D images, where scrolling is not possible, to see if subjects are capable of integrating multiple slices into a localization response identifying a target of interest.
A total of eight experimental conditions were evaluated (2D versus 3D images, large versus small targets, power-law versus white noise). The team evaluated performance in terms of task efficiency and classification image technique, which shows how observers use noisy images to perform visual tasks, such as target localization.
According to Abbey, “The somewhat surprising finding of the paper was that the classification images showed almost no evidence of combining information over multiple sections of the image to localize a target that spans multiple sections of the image. The observers are essentially treating the volumetric image as a stack of independent 2D images. This leads to a dissociation in which observers are more efficient at localizing larger targets in 2D images, and smaller targets (that don’t extend over many volumetric sections) in 3D images.”
The findings warrant further investigation, but they support and help to explain the need for multiple views in 3D image reading, and they provide useful information for modeling observer performance in volumetric images.
Read the open access research article by Abbey, Lago, and Eckstein, “Comparative observer effects in 2D and 3D localization tasks,” J. Medical Imaging 8(4), 041206 (2021), doi 10.1117/1.JMI.8.4.041206. The article is part of a JMI Special Series on 2D and 3D Imaging: Perspectives in Human and Model Observer Performance, guest edited by Claudia R. Mello-Thoms, Craig K. Abbey, and Elizabeth A. Krupinski.
Media Contact
Daneet Steffens
[email protected]
Original Source
https:/
Related Journal Article
http://dx.




