Machine learning study initiated at the Wyss Institute in collaboration with Google Research enables unprecedented AAV capsid diversification with potential for improving gene therapies
Credit: Wyss Institute at Harvard University (original by Drew Bryant)
(Boston) — Adeno-associated viruses (AAVs) have become promising vehicles for delivering gene therapies to defective tissues in the human body because they are non-pathogenic and can transfer therapeutic DNA into target cells. However, while the first gene therapy products approved by the Federal Drug Administration (FDA) use AAV vectors and others are likely to follow, AAV vectors still have not reached their full potential to meet gene therapeutic challenges.
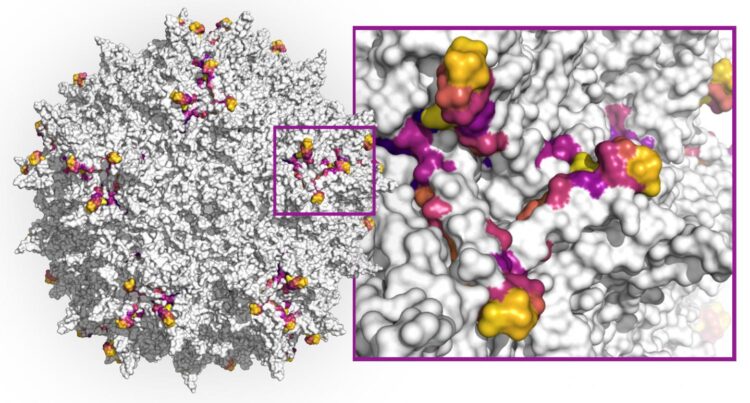
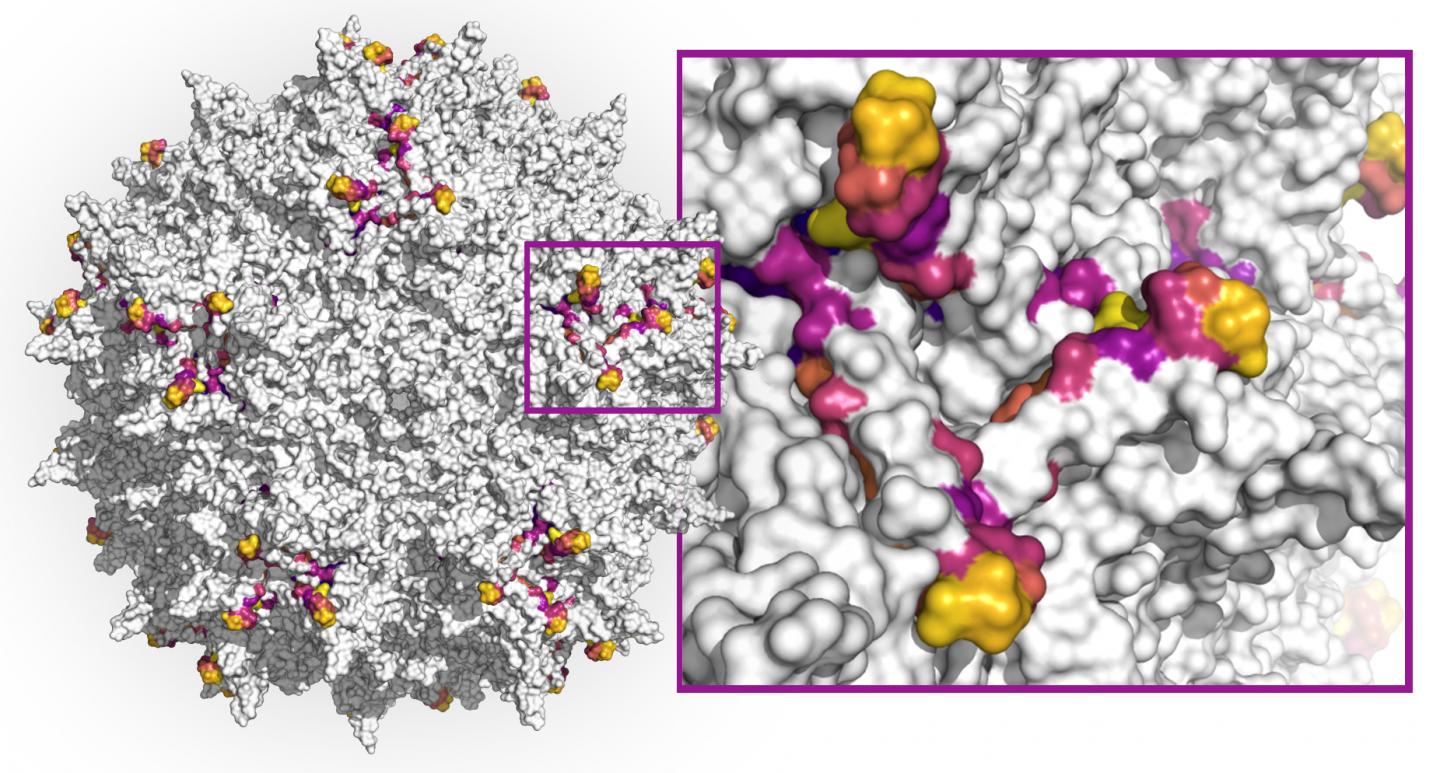
First, currently used AAV capsids – the spherical protein structures enveloping the virus’ single-stranded DNA genome which can be modified to encode therapeutic genes – are limited in their ability to specifically hone in on the tissue affected by a disease and their wider distribution throughout the human body causes them to be diluted. And secondly, patients’ immune systems, after having been exposed to a similar AAV virus, can produce neutralizing antibodies that, even at low levels, can destroy AAVs upon re-exposure (neutralization), blocking the delivery of their therapeutic DNA payloads.
To overcome this neutralization problem, researchers are engineering enhanced AAV capsids they hope to be able to evade the immune system. Currently used methods, including “directed evolution” strategies that fast-track the evolution of a protein in laboratory conditions, only can create a limited diversity of capsids with most of them still resembling the naturally occurring AAV variants known as serotypes. However, it remains difficult to generate sufficient diversity using this approach without losing other desired functions of the capsid, such as their stability or ability to bind to specific cell types.
Now, a new study initiated by Wyss Core Faculty member George Church’s Synthetic Biology team at Harvard’s Wyss Institute for Biologically Inspired Engineering, and driven by a collaboration with Google Research has applied a computational deep learning approach to design highly diverse capsid variants from the AAV2 serotype across DNA sequences encoding a key protein segment that plays a role in immune-recognition as well as infection of target tissues. AAV2 is the most-studied serotype and has been used in the first FDA approved gene therapy, to treat a blinding disease.
Starting from a relatively small collection of capsid data, the team trained multiple machine learning methods and used them to design 200,000 virus variants. 110,689 of these variants produced viable AAV viruses. Between any two naturally occurring AAV serotypes, 12 amino acids within this segment are expected to differ. The team’s effort produced more than 57,000 variants that exhibited much higher diversity than this, some containing up to 29 combined substituted or additionally inserted amino acids. The findings are published in Nature Biotechnology.
“Our approach achieves the highest functional diversity of any capsid library thus far. It unlocks vast areas of functional but previously unreachable sequence space, with many potential applications for generating improved viral vectors, like AAVs with much reduced immunogenicity and much improved target tissue selectivity, and also for highly efficient gene therapies,” said last-author Eric Kelsic, Ph.D., who started the project with Church, Ph.D., and co-founded the startup Dyno Therapeutics where he is now CEO. Dyno Therapeutics’ mission is to develop advanced gene therapy delivery vehicles by employing cutting-edge artificial intelligence (AI) approaches.
Using multiple design strategies, the team first generated smaller data sets on which they could train several machine learning models. These were collections of AAV capsids with variable numbers of mutations introduced in a 28 amino acid segment of the AAV2 VP3 protein that forms part of the capsid and exposes it to neutralizing antibodies. A high-throughput method enabling the synthesis of mutated capsid sequences and in vitro experiments for testing which ones efficiency produced viable stable capsids, provided a highly effective test bed for their overall approach. The results from this first experimental study then were used by the team as training data for three alternative machine learning models that generated much larger numbers of diverse capsid variants to be tested with a final validation experiment.
A central bottleneck in the creation of diverse AAV capsids and variants that can evade neutralization is the production of capsids that remain stable: most of the variants will fail to assemble into functional capsids or package their AAV genomes. “The deep neural network models that we deployed with our Google collaborators accurately predicted capsid viability across extremely diverse variants. Reaching this level of diversity in the capsid segment is an important milestone that we can build on to find immune-evading capsids for gene therapy,” said co-first author Sam Sinai, Ph.D., a former graduate student of Church who joined Kelsic’s team at the Wyss Institute and is a co-founder leading the machine learning team at Dyno Therapeutics. “And we can take similar approaches to create AAV capsids with much improved tissue selectivity.”
In 2019, a former Wyss team including Kelsic, Sinai, and their mentor Church published a related approach in Science in which they mutated one by one each of the 735 amino acids within the entire AAV2 capsid in different ways. What they called a “wide” search resulted in a large AAV library that identified changes affecting AAV2’s viability and its “homing” potential to specific organs in mice, as well as a previously unknown accessory protein that binds to cell membranes and which was hidden within the capsid-encoding DNA sequence. In their previous study, the researchers used a simple experimental model to optimize the tissue targeting ability of the virus.
“This new study involving machine learning models developed with Google Research nicely complements our earlier work in that it focuses on a small, but very important, region of the AAV capsid with an unprecedented resolution,” said co-corresponding author Church. “It shows that neural networks combined with the high-throughput synthetic testing developed in our lab is changing the way we design gene delivery vehicles and protein drugs.” Church is the lead of the Wyss Institute’s Synthetic Biology platform where the project was started, and Professor of Genetics at Harvard Medical School and of Health Sciences and Technology at Harvard and MIT.
“This work gives a glimpse into the future as artificial intelligence approaches, such as machine learning, are opening up vast new design spaces that enable the development of entirely new drugs and drug delivery approaches for combating innumerable challenges to human health. It also highlights the Wyss Institute’s commitment to computational problem-solving in areas where new therapies are desperately needed,” said Wyss Founding Director Donald Ingber, M.D., Ph.D., who is also the Judah Folkman Professor of Vascular Biology at Harvard Medical School and Boston Children’s Hospital, and Professor of Bioengineering at SEAS.
###
By Benjamin Boettner
Other authors on the study were co-first authors Drew H. Bryant and Ali Bashir, as well as Patrick F. Riley at Google Research; Nina K. Jain and Pierce J. Ogden at the Wyss Institute and HMS; and co-corresponding author Lucy J. Colwell at Google Research and the University of Cambridge, UK. It was funded by Harvard’s Wyss Institute for Biologically Inspired Engineering.
PRESS CONTACTS
Wyss Institute for Biologically Inspired Engineering at Harvard University
Benjamin Boettner, [email protected], +1 617-432-8232
The Wyss Institute for Biologically Inspired Engineering at Harvard University (http://wyss.
Media Contact
Benjamin Boettner
[email protected]