Penn team of microbiologists and pulmonologists scan genome databases and find a new abundant viral family associated with disease
Credit: Penn Medicine
PHILADELPHIA – Viruses, the most abundant biological entities on earth, are a scourge on humanity, causing both chronic infections and global pandemics that can kill millions. Yet, the true extent of viruses that infect humans remains completely unknown. Some newly discovered viruses are recognized because of the sudden appearance of a new disease, such as SARS in 2003, or even HIV/AIDS in the early 1980s. New techniques, however, now enable scientists to identify viruses by directly studying RNA or DNA sequences in genetic material associated with humans, enabling detection of whole populations of viruses – termed the virome – including those that may not cause acutely recognizable disease. However, identifying novel types of viruses is difficult as their genetic sequences may have little in common with already known viral genomes that are available in reference databases.
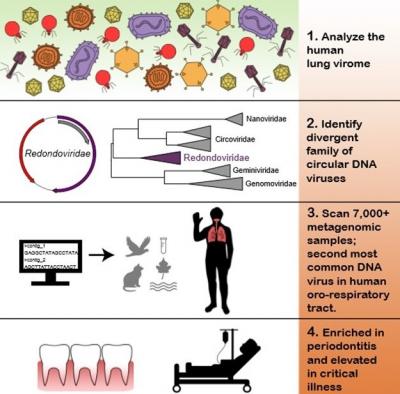
Researchers in the Perelman School of Medicine at the University of Pennsylvania, have now identified a previously unknown viral family, which turns out to be the second-most common DNA virus in human lung and mouth specimens, where it is associated with severe critical illness and gum disease, respectively. The team published their findings today in Cell Host Microbe. Senior authors Frederic D. Bushman, PhD, chair of the department of Microbiology, and Ronald G. Collman, MD, a professor of Pulmonary, Allergy and Critical Care, lead the team that discovered this new virus, which they dubbed Redondoviridae.
“New sequencing techniques have helped us uncover a world of new viruses,” said Bushman. “However, the majority of the sequence data we have so far remains unclassified, leaving us much work to do in order to better understand the human virome and how these new species may be associated with illness.”
The team analyzed samples from human lungs to sequence RNA and DNA in free-floating viral particles. Using this wide-ranging survey of viral sequences as a starting point to compare samples with known viral sequences in public databases, the team, including first authors Arwa A. Abbas and Louis J. Taylor, both graduate students working with Bushman and Collman, identified short sequences that were similar to a type of virus found in domesticated pig stool. From there, they identified whole genomes in the sample, which they recognized as a completely new human virus.
The team knew they might have discovered a virus because the sequence of DNA building blocks–that eventually form proteins–allowed them to recognize these proteins as distant relatives of known viral molecules, which are important for making the virus particle shell and managing replication.
With this new virus in hand, they then searched for it in additional DNA sequence data. After scanning more than 7,000 samples in databases, they found 17 complete redondovirus genomes, and many more with partial sequences. That search revealed that this family of small, circular DNA viruses is associated with periodontitis–inflammation of the gums. They searched for it directly in patient samples and discovered that redondoviruses were also particularly abundant in the lungs of critically ill patients in intensive care units (ICU).
“Overall, we are asking whether we can we take unknown DNA sequences and make sense of it by identifying new viruses from the whole universe of sequences in the human virome,” Collman said.
The team is now working to grow redondovirus in the lab in order to investigate basic questions about its biology, as well as more clinical questions about its relationship to diseases. They hope that this direction will help determine if the new virus is simply associated with disease or whether it causes disease, and physicians might be able to use this knowledge to better help patients in the ICU and with dental disease.
###
Additional Penn authors include Marisol I. Dothard, Jacob S. Leiby, Ayannah S. Fitzgerald, and Layla A. Khatib. This work was funded by the National Institutes of Health (R61-HL137063, R01-HL113252) and the National Science Foundation, (DGE-1321851, T32-AI- 17 007324).
Penn Medicine is one of the world’s leading academic medical centers, dedicated to the related missions of medical education, biomedical research, and excellence in patient care. Penn Medicine consists of the Raymond and Ruth Perelman School of Medicine at the University of Pennsylvania (founded in 1765 as the nation’s first medical school) and the University of Pennsylvania Health System, which together form a $7.8 billion enterprise.
The Perelman School of Medicine has been ranked among the top medical schools in the United States for more than 20 years, according to U.S. News & World Report’s survey of research-oriented medical schools. The School is consistently among the nation’s top recipients of funding from the National Institutes of Health, with $425 million awarded in the 2018 fiscal year.
The University of Pennsylvania Health System’s patient care facilities include: the Hospital of the University of Pennsylvania and Penn Presbyterian Medical Center–which are recognized as one of the nation’s top “Honor Roll” hospitals by U.S. News & World Report–Chester County Hospital; Lancaster General Health; Penn Medicine Princeton Health; and Pennsylvania Hospital, the nation’s first hospital, founded in 1751. Additional facilities and enterprises include Good Shepherd Penn Partners, Penn Home Care and Hospice Services, Lancaster Behavioral Health Hospital, and Princeton House Behavioral Health, among others.
Penn Medicine is powered by a talented and dedicated workforce of more than 40,000 people. The organization also has alliances with top community health systems across both Southeastern Pennsylvania and Southern New Jersey, creating more options for patients no matter where they live.
Penn Medicine is committed to improving lives and health through a variety of community-based programs and activities. In fiscal year 2018, Penn Medicine provided more than $525 million to benefit our community.
Media Contact
Karen Kreeger
[email protected]