Credit: NIAID
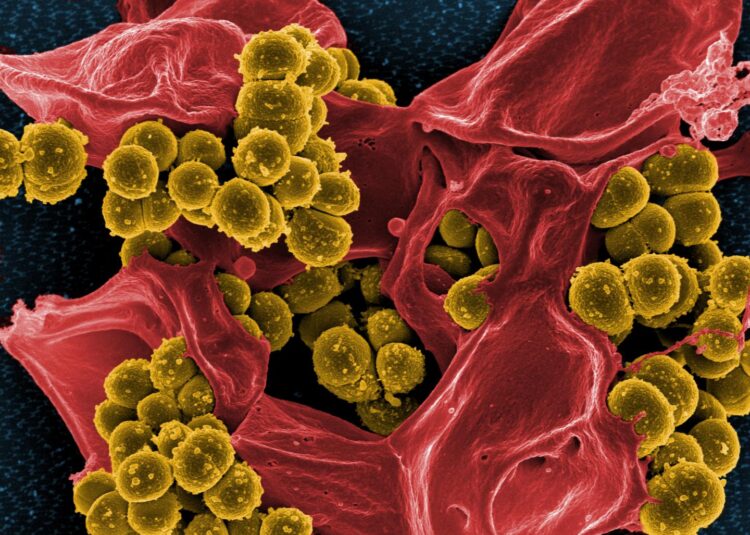
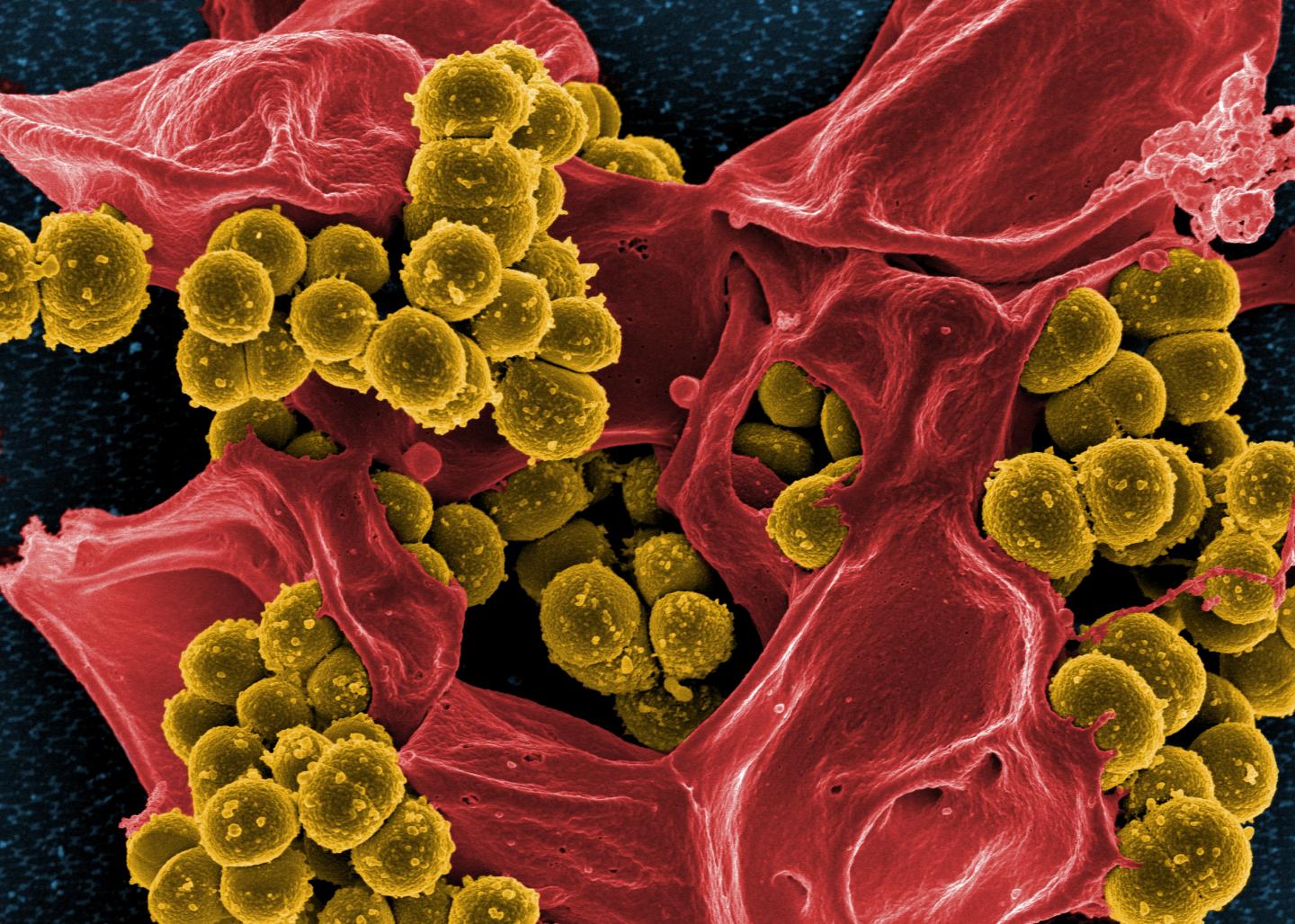
The bacterium Staphylococcus epidermidisis primarily a harmless microbe found on the skin and in the noses of humans. Yet some strains of this species can cause infections – in catheters, artificial joints, heart valves, and in the bloodstream – which are difficult to treat. These bacteria are often resistant to a particularly effective antibiotic, methicillin, and are among the most feared germs in hospitals. How these usually harmless skin microbes become deadly pathogens has been unclear up to now.
An international research team has now discovered what distinguishes peaceful S. epidermidis microorganisms from the many dangerous invaders. The scientists have identified a new gene cluster that enables the more aggressive bacteria to produce additional structures in their cell walls. This morphological alteration allows the staphylococci to attach more easily to human cells forming the blood vessels, a process via which they can persist in the bloodstream to become pathogens. These new cell wall structures may also allow the spread of methicillin resistance, by transferring it, for example, from Staphylococcus epidermidis to its more dangerous relative Staphylococcus aureus.
The study was carried out under the direction of researchers of the Cluster of Excellence “Controlling Microbes to Fight Infections” (CMFI) of the University of Tübingen and the German Center for Infection Research (DZIF) in cooperation with universities in Copenhagen, Hamburg, Shanghai and Hanover as well as the German Center for Lung Research (DZL) in Borstel. The results are being published in the journal Nature Microbiology.
Set apart by structure
A considerable portion of the cell walls of Staphylococci – like other gram-positive bacteria – is made up of teichoic acids. Chain-like, these polymers cover the bacterial surface. Their chemical structures vary according to species. “During our examination we determined that many pathogenic strains of S. epidermidis have an additional gene cluster that contains information for the synthesis of wall teichoic acids that are actually typical of S. aureus,” says researcher Dr. Xin Du of the Cluster of Excellence of the CMFI and DZIF. She adds that experiments have shown S. epidermidis bacteria with only species-specific teichoic acids in their walls are not very invasive, colonizing the surfaces of the skin and mucous membranes. If the wall teichoic acids for S. aureus are also present, Xin Du explains, they are unable to attach effectively to those surfaces. Instead, they are more successful in penetrating the tissues of their human host. “At some point, a few S. epidermidis clones took on the corresponding genes from S. aureus and became threatening pathogens as a result,” says Professor Andreas Peschel of the Cluster of Excellence CMFI and of the DZIF.
It’s long been known that bacteria can share genetic material through gene transfer. Bacteriophages – viruses that infect bacteria – carry out the transfer. Mostly, this takes place within one species and requires similar surface structures to which the bacteriophages bind. “Differing cell wall structures normally prevent gene transfer between S. epidermidis and S. aureus. But in S. epidermidis strains that can also produce the wall teichoic acids of S. aureus, that type of gene transfer suddenly becomes possible between different species,” explains Peschel. That would explain, he continues, how S. epidermidis could transfer methicillin resistance to even more threatening – and then methicillin-resistant – S. aureus, adding that more investigation is still needed. The new findings are an important step, says Peschel, towards developing better treatments or vaccinations against dangerous pathogens such as S. epidermidis ST 23, which has been known for fifteen years and belongs to the group of HA-MRSE (healthcare-associated methicillin-resistant S. epidermidis).
###
Media Contact
Prof. Dr. Andreas Peschel
[email protected]
Original Source
https:/
Related Journal Article
http://dx.