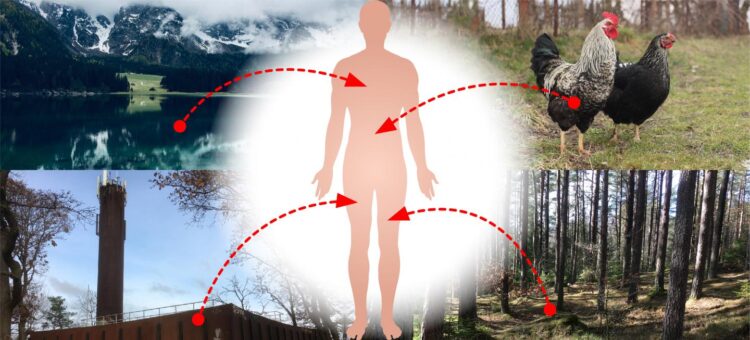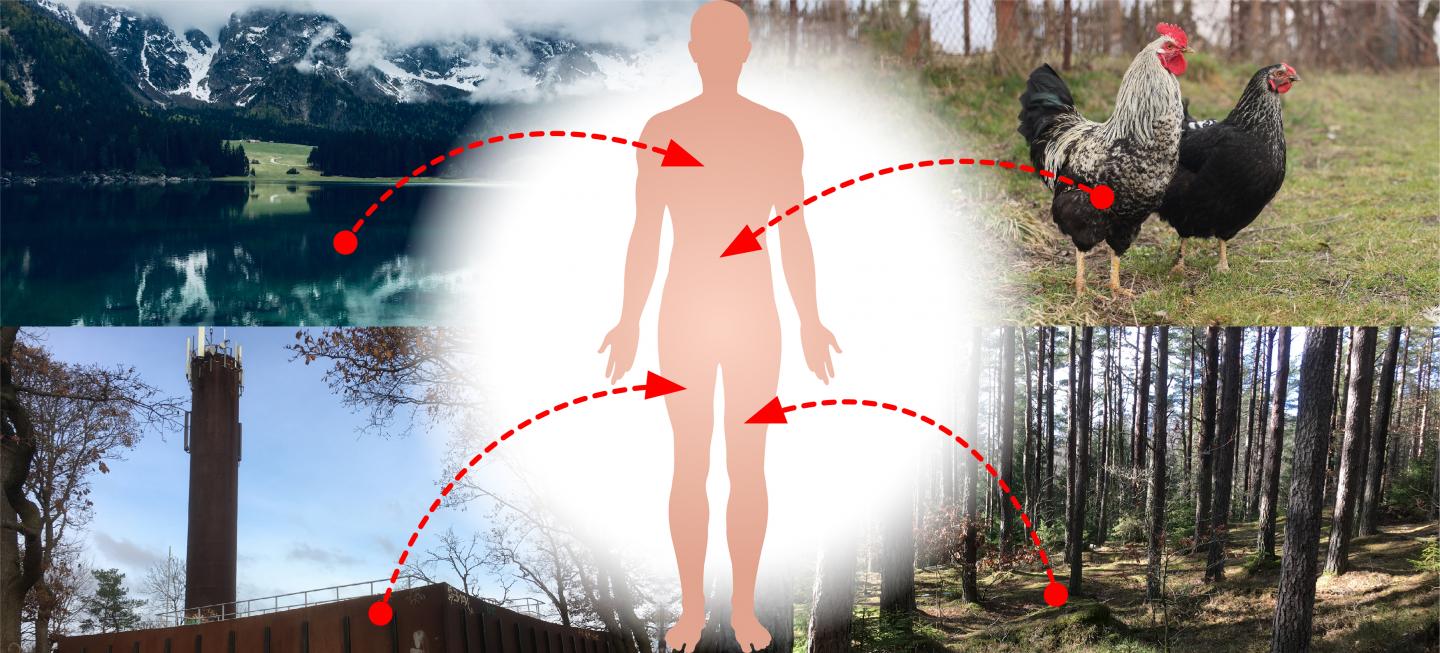
Credit: Jan Zrimec/Chalmers University of Technology
Pathogenic bacteria in humans are developing resistance to antibiotics much faster than expected. Now, computational research at Chalmers University of Technology, Sweden, shows that one reason could be significant genetic transfer between bacteria in our ecosystems and to humans. This work has also led to new tools for resistance researchers.
According to the World Health Organisation, antibiotic resistance is one of the greatest threats to global health, food safety and development. It already causes over 33,000 deaths a year in Europe alone.
Completely different species of bacteria can spread resistance genes to each other through plasmids – small DNA molecules where bacteria store some of their genes outside the chromosome. When two bacterial cells come into contact, they can copy plasmids to each other. This is called conjugation, and it is the most important mechanism for spreading antibiotic resistance.
“In recent years, we’ve seen that resistance genes spread to human pathogens to a much greater degree than anyone expected,” says Jan Zrimec, researcher in systems and synthetic biology at Chalmers University of Technology. “Many of the genes appear to have originated in a wide array of bacterial species and environments, such as soil, water and plant bacteria.
“This has been difficult to explain, because although conjugation is very common, we’ve thought that there was a distinct limitation for which bacterial species can transfer plasmids to each other. Plasmids belong to different mobility groups, or MOB groups, so they can’t transfer between just any bacterial species.
Zrimec has developed new methods of data analysis that show that genetic transfer may be much more boundless and widespread than previously expected.
Among other things, he has developed an algorithm that can identify specific DNA regions that are necessary for conjugation – called oriT regions – in large amounts of data consisting of genetic sequences from the DNA of thousands of plasmids. The algorithm can also sort plasmids into MOB groups based on the identified oriT regions.
He has used the algorithm to explore known gene sequences from over 4,600 naturally occurring plasmids from different types of bacteria, which has not been possible to do systematically before. The results show, among other things, that:
- The number of oriT regions may be almost eight times higher than those found with the standard method used today.
- The number of mobile plasmids may be twice as high as previously known.
- The number of bacterial species that have mobile plasmids may be almost twice as high as previously known.
- Over half of these plasmids have oriT regions that match a conjugation enzyme from another plasmid that has previously been classified in a different MOB group. This means that they could be transferred by one of these plasmids that happens to be in the same bacterial cell.
The last part means that there may be transfer mechanisms between large numbers of bacterial species and environments where we previously believed there were barriers.
“These results could imply that there is a robust network for transferring plasmids between bacteria in humans, animals, plants, soil, aquatic environments and industries, to name a few,” Zrimec explains. “Resistance genes occur naturally in many different bacteria in these ecosystems, and the hypothetical network could mean that genes from all of these environments can be transferred to bacteria that cause disease in humans.
“This may be a possible reason for the rapid development of resistance in human pathogens that we have observed in recent years. Our extensive use of antibiotics selects for resistance genes, which could thus flow in from a much larger naturally occurring genetic reservoir than we previously estimated.”
The results need to be verified experimentally in the future, but the data analysis methods Zrimec developed can already be employed by many of the researchers working with antibiotic resistance in various medical and biological fields. They provide a powerful new tool for systematically mapping out the potential transferability of different plasmids.
“This has been a major limitation of the research field up to now,” Zrimec says. “I hope that the methods will be able to benefit large parts of the research into antibiotic resistance, which is an extremely interdisciplinary and fragmented area. The methods can be used for studies aiming to develop more effective limitations to antibiotic use, instructions for how antibiotics are to be used and new types of substances that can prevent the spread of resistance genes at the molecular level.”
More about: Genetic transfer through conjugation:
In order for conjugation to start, an enzyme is needed – a relaxase – which fits onto a specific location on the plasmid. The relaxase has to recognise and bind to a region where the DNA ring can be nicked and a strand can be transferred to the next bacterium. This DNA region is called the origin of transfer, or oriT.
Previously, it was thought that an individual plasmid must contain both the gene for the relaxase and a matching oriT in order to be transferred to other bacteria. But a bacterial cell can contain several plasmids, and in recent years various researchers have shown that a relaxase from one plasmid can fit with an oriT region on another plasmid in the same cell and activate the conjugation of that plasmid.
This means that it may be enough for a plasmid to have only an oriT to be able to conjugate, which in turn means that many plasmids that have previously been classed as nonmobile, because they lack the relaxase gene, can be conjugative. But until now it has not been known how common the phenomenon is among bacteria. This is one of the knowledge gaps that Zrimec’s results are helping to fill in.
More about: The new method compared with the current standard:
The current standard tools for assessing the transferability of plasmids are based on searching for the DNA sequences for the relaxase enzyme or for oriT regions that the enzyme can bind to. There are several key limitations to this. For one, some tools produce incomplete results, while others require extremely time-consuming and resource-demanding laboratory tests.
Zrimec’s new data analysis method is based solely on identifying oriT regions, using special physiochemical properties found specifically in oriT regions of DNA. Through previous research, he has shown that these physiochemical signatures – which determine which relaxase can bind to the oriT region – are more stable and specific than the DNA sequences themselves. This allows the classification of the plasmids to the right MOB group based on the oriT region, independently of relaxase, which also allows researchers to map out the overall transferability between different bacterial species and environments.
The method can manage large amounts of data and can be used to search effectively for oriT regions on plasmids in their entirety.
###
Media Contact
Johanna Wilde
[email protected]
Original Source
https:/
Related Journal Article
http://dx.