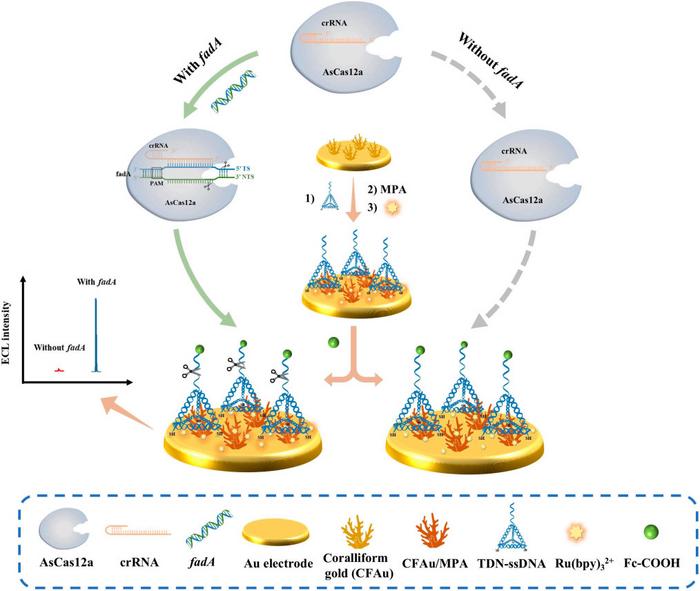
In a groundbreaking advancement in the field of diagnostic biosensing, researchers have engineered a sophisticated amplification-free electrochemical biosensor designed to detect Fusobacterium nucleatum. This specific bacterium has garnered attention due to its significant correlation with colorectal cancer. The biosensor exploits the unique properties of the CRISPR/Cas12a system, known for its high selectivity and remarkable nucleic acid cleavage capabilities, creating a promising tool for early diagnosis of cancerous conditions.
The underlying mechanism of this innovative sensor lies in its strategic integration of tetrahedral DNA nanostructures (TDNs) and coralliform gold (CFAu) nanostructures. The TDNs serve as a scaffold that significantly enhances the recognition and cleavage efficiency of the Cas12a enzyme, thereby amplifying the biosensor’s responsiveness in the presence of target nucleotides. The incorporation of these elements not only provides a unique structural advantage but also addresses common challenges faced by traditional CRISPR-based sensors, such as probe aggregation or entanglement, which can diminish enzyme efficacy.
Electrochemiluminescence (ECL) has been identified as a vital component in this biosensing platform. ECL is heralded for its sensitivity, allowing for the reliable detection of minute quantities of target nucleic acids, proteins, and small molecules. As the luminescent reactants on the electrode are regenerated through precise electrochemical reactions, the sensor benefits from enhanced photon production during measurement cycles. This characteristic is crucial, enabling heightened sensitivity and specificity when detecting F. nucleatum, particularly given the complexities associated with diagnosing infections caused by this pathogen.
.adsslot_wUQn36cGlE{width:728px !important;height:90px !important;}
@media(max-width:1199px){ .adsslot_wUQn36cGlE{width:468px !important;height:60px !important;}
}
@media(max-width:767px){ .adsslot_wUQn36cGlE{width:320px !important;height:50px !important;}
}
ADVERTISEMENT
A key aspect of the newly developed biosensor is its ability to operate within an amplification-free framework. Unlike conventional methods reliant on exponential amplification techniques, this sensor achieves remarkable detection limits down to one colony-forming unit per milliliter, demonstrating its unparalleled capability in identifying low concentrations of the target bacterium. Such sensitivity not only highlights the practicality of the biosensor in clinical settings but also sets a precedent for future innovations in biosensor design.
The process of biosensor construction involved meticulous electrochemical deposition of CFAu nanostructures, providing an ideal surface for immobilizing TDN-ssDNA through sulfur-gold bonding. In a subsequent step, a self-assembled monolayer of 3-Mercaptopropionic acid (MPA) was created. The deliberate choice of materials enhances the surface chemistry of the sensor, paving the way for effective coupling with luminescent agents, such as ruthenium tris(bipyridine) [Ru(bpy)3^2+].
Significantly, the biosensor’s performance is directly contingent upon the specific interactions between the TDNs, the Cas12a enzyme, and the target ‘fadA’ gene present in F. nucleatum. In the presence of the ‘fadA’ gene, the AsCas12a enzyme exhibits trans-cleavage activity, leading to the cleavage of fluorescent probes. This reaction triggers an increase in electrochemiluminescent signals, providing a clear indication of the presence of F. nucleatum. Conversely, in the absence of the target gene, the sensor’s signal remains almost undetectable, underscoring its specificity.
One of the compelling advantages of this biosensor is its adaptability. By rational design of CRISPR RNA (crRNA) sequences, it can be tailored for the detection of a wide range of nucleic acids and pathogens. This versatility positions the biosensor as an invaluable tool not only for detecting F. nucleatum but also for broader applications including the diagnosis of other bacterial infections and various diseases. The implications of this flexibility are vast, providing a pathway for future research and diagnostic innovations.
Furthermore, the study reveals that this biosensing technology is bolstered by its exceptional linear detection range, spanning from 10 femtomoles to 100 nanomoles. This characteristic, combined with its high mismatch sensitivity, allows the biosensor to differentiate between wild-type sequences and mutations. This feature is particularly advantageous for clinical diagnostics, where distinguishing between similar nucleic acid sequences can be crucial for accurate diagnosis and treatment pathways.
The research team, led by Jieling Qin of the Beijing Institute of Technology, emphasizes the significance of their findings. The implications of this biosensor could revolutionize how medical professionals diagnose infectious diseases, specifically in cases where early detection is vital to successful treatment outcomes. By streamlining the detection process and enhancing efficiency, the biosensor has the potential to expedite diagnostics, ultimately improving patient care.
Support for this innovative research comes from various funding bodies, including the China Postdoctoral Science Foundation and the Beijing Institute of Technology Research Fund Program for Young Scholars, which underscores the collaborative efforts aimed at tackling pressing healthcare challenges through technological advancements. The collective aspiration is to refine diagnostic tools and pave the way for future innovations that could have a lasting impact on health outcomes globally.
The results of this significant study have been documented in the recent publication titled “Amplification-Free Electrochemiluminescent Biosensor for Ultrasensitive Detection of Fusobacterium nucleatum Using Tetrahedral DNA-Based CRISPR/Cas12a,” which appeared in the journal Cyborg and Bionic Systems on May 1, 2025. This dissemination of knowledge not only highlights the advancements made in the field of biosensing but also sparks a conversation about the future of molecular diagnostics in the fight against cancer and infectious diseases.
As the world grapples with a myriad of health challenges, such developments in biosensing technologies are crucial. With the ability to detect specific pathogens efficiently and accurately, researchers and healthcare professionals are better equipped to make informed decisions that could save lives. The ongoing evolution of CRISPR technology combined with innovative engineering and biochemistry efforts marks a pivotal moment in the history of disease detection, fostering hope for a future marked by enhanced diagnostics and improved healthcare systems.
In conclusion, this research showcases an exceptional leap forward in the integration of advanced biotechnology and nanostructured materials to create a powerful diagnostic tool. As the scientific community continues to explore the possibilities that reside within CRISPR technology and biosensing frameworks, it is clear that the future holds exciting potential for the early detection and diagnosis of diseases, promising more effective intervention strategies.
Subject of Research: Electrochemical biosensor for detecting Fusobacterium nucleatum
Article Title: Amplification-Free Electrochemiluminescent Biosensor for Ultrasensitive Detection of Fusobacterium nucleatum Using Tetrahedral DNA-Based CRISPR/Cas12a
News Publication Date: May 1, 2025
Web References: Not provided
References: Not provided
Image Credits: Jieling Qin, School of Chemistry and Chemical Engineering, Beijing Institute of Technology Zhengzhou Academy of Intelligent Technology, Beijing Institute of Technology
Keywords
Health and medicine, Applied sciences and engineering, Life sciences
Tags: Advanced Diagnostic Toolsamplification-free biosensing methodscolorectal cancer biomarkersCRISPR-Cas12a technologyearly diagnosis of cancerelectrochemical biosensor for cancer detectionelectrochemiluminescence in diagnosticsFusobacterium nucleatum detectionnanostructured biosensorsselective nucleic acid detectiontetrahedral DNA nanostructuresultra-sensitive biosensing techniques