Scientists are starting to understand the precise workings of a type of gene that, unlike other genes, does not code for proteins – the building blocks of life.
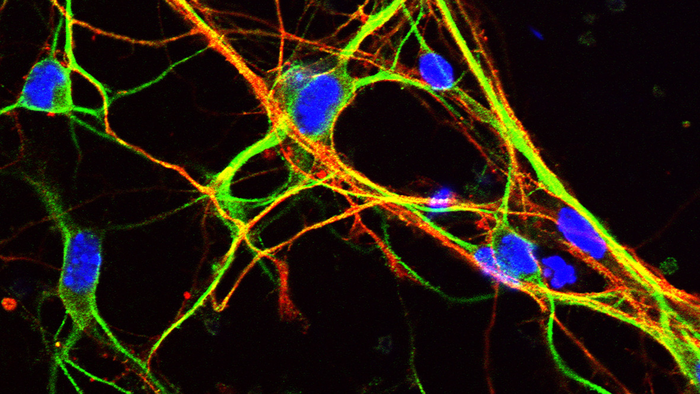
Credit: Dr Robert Williams, University of Bath
Scientists are starting to understand the precise workings of a type of gene that, unlike other genes, does not code for proteins – the building blocks of life.
New research led by the University of Bath shows the mechanism by which genes coding for a subset of long non-coding RNA (lncRNA) interact with neighbouring genes to regulate the development and function of essential nerve cells.
Despite their prevalence on genes coding for lncRNA in the genome (estimates range from 18,000-60,000 lncRNA genes in the human genome compared to 20,000 protein-coding genes), these segments of DNA were once written-off as junk precisely because the information contained within them does not result in the production of a protein. However, it is now clear that some lncRNAs are anything but scrap, and these may come to play a key role in restoring physical function in people who have suffered serious nerve damage.
Although the function of most lncRNA genes remains a mystery, a subset are co-expressed in the brain along with neighbouring genes that code for proteins involved in gene expression control. In other words, genes for these lncRNAs and their protein-coding neighbours work as a pair. Together, they regulate the development and function of essential nerve cells, particularly in the brain during embryonic development and in early life.
The new study describes the regulatory pathway involved in controlling the levels of one of these gene pairs. Their location and quantity in the genome need to be carefully coordinated, as does the timing of their activity.
“We previously defined one of the most profound functions for lncRNA in the brain and our new study identifies an important signalling pathway that acts to coordinate the expression of this lncRNA and the key protein coding gene that it is paired with,” explains Dr Keith Vance, lead author of the study from the Department of Biology & Biochemistry at Bath.
“This new research takes us closer to understanding the basic biology of nerve cells and how they are produced. Regenerative medicine is the end-game and with further research we hope to develop a deeper understanding of how lncRNA genes operate in the brain.”
He adds: “This knowledge could be important for scientists looking for ways to replace defective neurons and restore nerve function – for instance in people who have had strokes.”
The research was funded by the Biotechnology and Biological Sciences Research Council (BBSRC) and is published today in PLOS Genetics.
Journal
PLoS Genetics
DOI
10.1371/journal.pgen.1010230
Method of Research
Experimental study
Subject of Research
People
Article Title
Chromatin interaction maps identify Wnt responsive cis-regulatory elements coordinating Paupar-Pax6 expression in neuronal cells
Article Publication Date
16-Jun-2022




