Clinical studies of COVID-19 drugs may benefit from randomization and early recruitment, model shows
Credit: (c) Tomoki Shimizu / Kyoto University
As scientists continue to search high and low for effective COVID-19 treatments, a new modeling study suggests that randomization, early patient enrollment and treatment initiation in clinical trials could be the keys to identifying effective antiviral drugs.
The researchers–led by Shingo Iwami, associate investigator at the Kyoto University Institute for the Advanced Study of Human Biology (ASHBi) and Keisuke Ejima, assistant research scientist at Indiana University Bloomington–report their findings in PLOS Medicine.
“Almost all antiviral clinical trials have failed to observe a significant effect against SARS-CoV-2, so we wanted to show why and what is important for an optimal study design,” shares first author Shoya Iwanami, an assistant professor at Nagoya University.
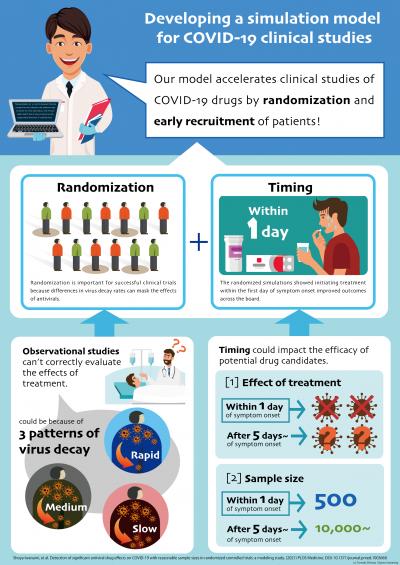
Given the inconsistent results of past trials, Iwanami and colleagues used a mathematical model to first analyze longitudinal patient data from clinical research. By simulating the amount of virus in the upper respiratory specimens, the team found that virus-producing cells died at different rates, classifying patients into those with rapid, medium or slow virus decay.
In observational studies, physicians assess whether and when patients should receive antiviral treatment based on their symptoms, as opposed to randomization where patients are blindly assigned to treatment and control groups. Since slow decay may be associated with more severe disease, observational studies may have been limited to certain patients, failing to capture the spectrum of viral dynamics and confounding the results.
“We found that for successful clinical trials, randomization is important because [differences in virus decay rates] can affect the effects of antivirals, ” Iwanami explains.
Aside from randomization, the researchers also found that timing could impact the efficacy of potential drug candidates. Regardless of virus decay group, their simulations showed that initiating treatment five days after symptom onset masked drug efficacy. Meanwhile, administering antivirals within the first day of symptom onset improved outcomes across the board.
To mimic randomized controlled trials–considered the gold standard for evaluating interventions–the researchers added hypothetical drugs with high SARS-CoV-2 inhibition rates into their model. As patients in past studies were often recruited without considering treatment timing, the team’s model revealed that these clinical trials would have needed to enroll over 10,000 participants per group to provide statistically significant data on drug efficacy–a practical challenge in terms of patient recruitment and resource availability.
In contrast, if patients are recruited early and treated within a day of symptom onset, the team found that the required sample size would drop to just 584 participants per group for an antiviral with 95 percent inhibition, and 458 per group for 99 percent inhibition. Ultimately, their findings highlight the importance of randomization as well as prompt patient recruitment and treatment initiation in evaluating COVID-19 drug candidates.
“We could apply this process to other clinical trials or diseases. This model on clinical design can accelerate drug repositioning or new drug development, ” says Iwanami, adding that trials following these recommendations are now underway.
###
Paper Information:
Shoya Iwanami, Keisuke Ejima*, Kwang Su Kim, Koji Noshita, Yasuhisa Fujita, Taiga Miyazaki, Shigeru Kohno, Yoshitsugu Miyazaki, Shimpei Morimoto, Shinji Nakaoka, Yoshiki Koizumi, Yusuke Asai, Kazuyuki Aihara, Koichi Watashi, Robin N. Thompson, Kenji Shibuya, Katsuhito Fujiu, Alan S. Perelson, Shingo Iwami*, and Takaji Wakita (2021). Detection of significant antiviral drug effects on COVID-19 with reasonable sample sizes in randomized controlled trials: a modeling study, PLOS Medicine, DOI: 10.1371/journal.pmed.1003660.
*Co-corresponding authors
Media Contact
Tomoki Shimizu
[email protected]
Related Journal Article
http://dx.