Study underscores the need for greatly increased genome surveillance to rapidly identify and track the emergence and introduction of SARS-CoV-2 variants in the United States, investigators report in The American Journal of Pathology
Credit: Houston Methodist
Philadelphia, April 14, 2021 – In late 2020, several concerning SARS-CoV-2 variants emerged globally. They are believed to be more easily transmissible, and there is concern that some may reduce the effectiveness of antibody treatments and vaccines. An extensive genome sequencing program run by the Houston Methodist health system has identified all six of the currently identified SARS-CoV-2 variants in their patients. A new study appearing in The American Journal of Pathology, published by Elsevier, finds that the variants are widely spread across the Houston metropolitan area.
“Before the SARS-CoV-2 virus arrived in Houston, we planned an integrated strategy to confront and mitigate this microbial threat to our patients,” explains lead investigator James M. Musser, MD, PhD, chair of the Department of Pathology and Genomic Medicine at Houston Methodist and Fondren Presidential Distinguished Chair with Houston Methodist Research Institute, Houston, TX, USA. “We instituted a plan to sequence the genome of every positive specimen from patients within the Houston Methodist system with the goal of understanding pathogen spread in our community and identifying biologically-important mutant viruses.”
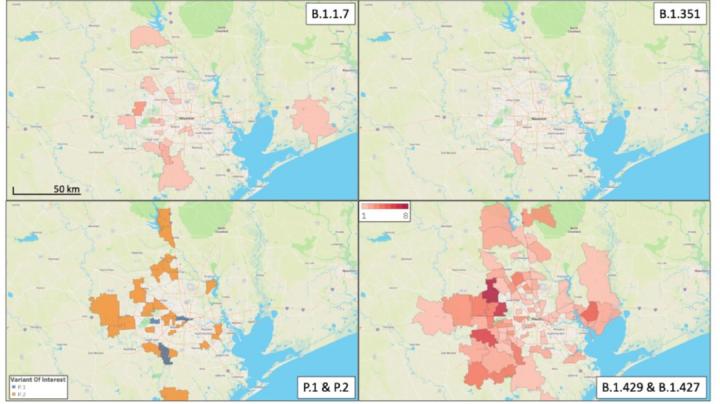
Dr. Musser notes that Greater Houston is the first metropolitan multiplex in the United States that has documented all of these important and concerning variants circulating among its residents. The Houston Methodist health system has one of the largest SARS-CoV-2 genome sequencing operations in the US and has so far sequenced 20,453 genomes from clinical specimens collected from patients from March 2020 to February 2021. Patient home address zip codes were used to see how the variants spread across Greater Houston’s large, multiethnic, and geographically and socioeconomically diverse population.
The UK variant known as B.1.1.7 was first identified in a patient in Houston in January 2021 and has been found in 23 patients through February 2021. The California variants, B.1.429 and B.1.427 were first found in Houston in late December 2020. Variant B.1.429 has been identified in 143 patients in Houston and variant B.1.427 in 19 patients. The South African variant B.1.351 was first detected in Houston in two patients in December 2020 and January 2021. The Brazil variant P.1 has been identified in four patients since mid-January 2021 and P.2 in 39 patients since late December 2020.
With the exception of B.1.351, the investigators found geographically widespread dissemination of the variants, indicating successful patient-to-patient transmission among the communities. None of the patients were from a common household or reported recent international travel, suggesting that every infection was independently acquired locally or during domestic travel.
Initially Dr. Musser and co-investigators had observed that although the P.2 infection was numerically a minor cause of all Houston-area infections, it was the most common variant of concern in their population because its mutation has the potential to neutralize treatments and vaccines. Continued monitoring of the Houston area shows that the UK B.1.1.7 variant is now the most prevalent variant. According to Dr. Musser, “Emergence of new variants underscores the need for ongoing extensive genomic sequencing efforts for early identification and public health warning.”
By linking the SARS-CoV-2 whole genome sequencing data to patient metadata in the electronic medical record, Houston Methodist is able to use analytic tools such as high-performance compute clusters and machine learning to investigate the genomic diversity and characteristics such as strain virulence or patient outcomes. “Our goal is to sequence the SARS-CoV-2 genome of every infected patient in our healthcare system in near-real time and expand outward to other patients in our community,” Dr. Musser states. “Our results underscore the need to greatly increase genome surveillance to rapidly identify and track the introduction of new SARS-CoV-2 variants in the U.S. and in local areas.”
Dr. Musser and his co-investigators have noted that in the last three weeks, Houston Methodist cases of the UK variant have increased to almost 1,600 from 648. They advise that because vaccination is the best way to decrease the number of circulating COVID-19 variants, a much larger percentage of the population needs to get vaccinated. In the meantime, the team is keeping a very close eye on the variants and the type of disease they cause.
###
Media Contact
Eileen Leahy
[email protected]
Related Journal Article
http://dx.