Credit: Sur Lab/MIT Picower Institute
To understand the massive capabilities and complexities of the brain, neuroscientists segment it into regions based on what they appear to do–like processing what we sense or how to move. What’s been lacking, however, is an ability to tie those functional maps precisely and consistently to matching distinctions of physical structure, especially in live animals while they are performing the functions of interest. In a new study, MIT researchers demonstrate a new way to do that, providing an unprecedented pairing of functional mapping in live mice with distinguishing structural information for each region all the way through the cortex into deeper tissue below.
“Our study shows for the first time that structural and functional coupling of visual areas in the mouse brain can be detected at sub-cellular resolution in vivo,” wrote the authors based in the lab of Mriganka Sur, Newton Professor of Neuroscience in The Picower Institute for Learning and Memory and the Department of Brain and Cognitive Sciences at MIT.
The technique could give scientists more precise ways to distinguish the borders and contents of regions they wish to study and could help them better understand the way that structural distinctions develop within individuals in different functional regions over time. Sur’s lab, for instance, is intensely interested in understanding the especially complex development of vision. Humans have 35 or so distinct functional regions that contribute to processing vision, Sur notes, and even mice have 10.
“There is something profound in the way that vision is represented and created in mammalian brains,” Sur said. “Where do these areas come from, what do they mean and what do they do? It has not been easy to understand how they differ. The critical thing is to precisely map or match the functional representation of each area with its anatomical uniqueness.”
Combining function and structure
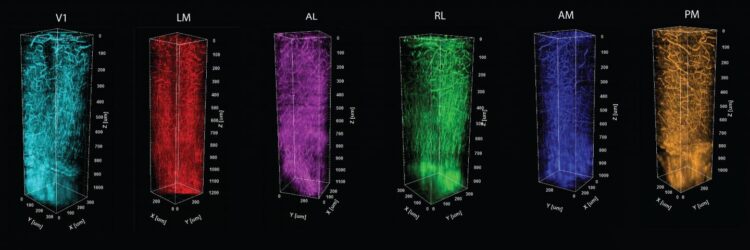
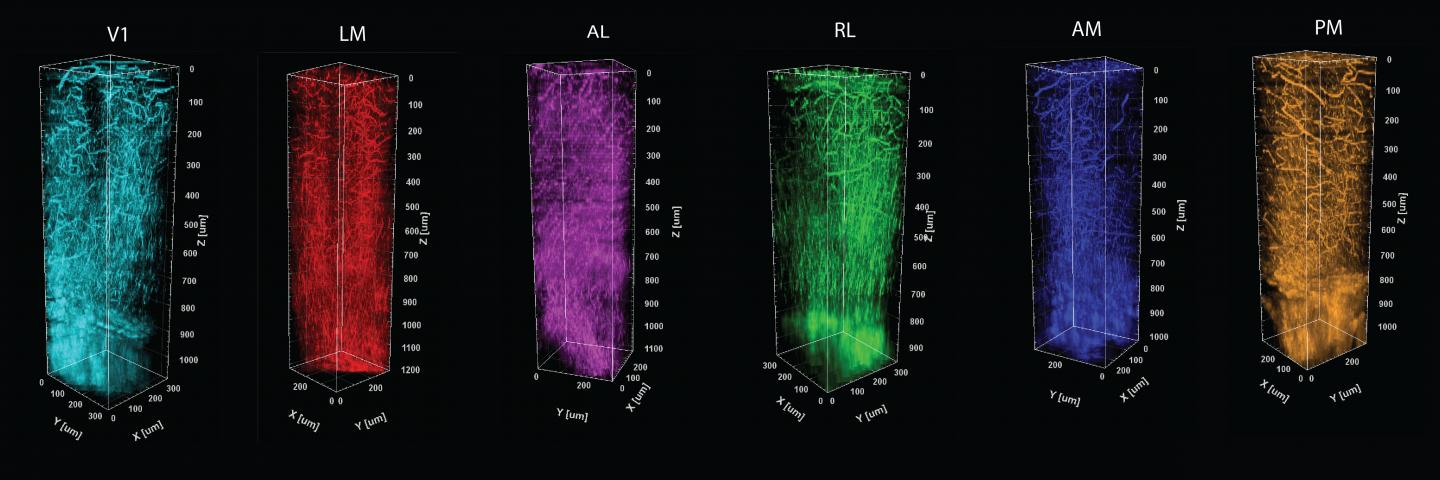
To develop tools to help answer those questions, postdoc Murat Yildirim led the study published in Biomedical Optics Express. In it he describes how the research team combined a method of charting functional areas–retinotopic mapping–with deep structural information measured by a technology he has helped to pioneer–third-harmonic generation (THG) three-photon microscopy.
In retinotopic mapping, researchers can identify functional regions by engineering neurons to flash when they become electrically active (and show changes in calcium) in response to a particular stimulation. For example, scientists could show a mouse a pattern moving across a screen and mark where neurons light up, with each area showing a characteristic location and pattern of response.
Three-photon microscopy can finely resolve individual cells and their smaller substructures as deep as a millimeter or more–enough to see all the way through the cortex. THG, meanwhile, adds the capability to finely resolve both blood vessels and the fibers of a material called myelin that wrap the long, tendrilous axons of many neurons. THG does not require adding any labeling dyes or chemicals.
Crucially, THG yields an important optical measure called effective attenuation length (EAL), which is a measure of how much the light is absorbed or scattered as it moves through the tissue. In the study, Yildirim and co-authors show that EAL specifically depends on each region’s unique architecture of cells, blood vessels and myelin. They measured EAL in each of six visual functional regions and showed that the EAL significantly differed among neighboring visual areas, providing a structural signature of sorts for each functional area. Their measurements were so precise, in fact, that they could show how EAL varied within functional regions, being most unique toward the middle and blending closer to the values of neighboring regions out toward the borders.
In other words, by combining the retinotopic mapping with THG three-photon microscopy, Yildirim said, scientists can identify distinct regions by both their function and structure while continuing to work with animals in live experiments. This can produce more accurate and faster results than making observations during behavior and then dissecting tissue in hopes of relocating those same exact positions in preserved brain sections later.
“We would like to combine the strength of retinotopic mapping with three-photon imaging to get more structural information,” Yildirim said. “Otherwise there may be some discrepancies when you do the live imaging of brain activity but then take the tissue out, stain it and try to find the same region.”
Especially as three-photon microscopy gains wider adoption and imaging speeds improve–right now imaging a millimeter deep column of cortex takes about 15 minutes, the authors acknowledge–the team expects its new method could be used not only for studies of the visual system but also in regions all around the cortex. Moreover it may help characterize disease states as well as healthy brain structure and function.
“This advance should enable similar studies of structural and functional coupling in other sensory and non-sensory cortical areas in the brains of mice and other animal models,” they wrote. “We believe that the structural and functional correlation in visual areas that we describe for the first time points to crucial developmental mechanisms that set up these areas, thus our work would lead to a better fundamental understanding of brain development, and of disorders such as Alzheimer’s, stroke and aging.”
###
In addition to Yildirim and Sur, the paper’s other authors are Ming Hu, Nhat Le, Hiroki Sugihara and Peter So.
The National Institutes of Health, the National Science Foundation, The JPB Foundation and the Massachusetts Life Sciences Initiative provided funding for the study.
Media Contact
David Orenstein
[email protected]
Original Source
http://picower.
Related Journal Article
http://dx.