Shipwreck harbors a rich spatially structured microbial community
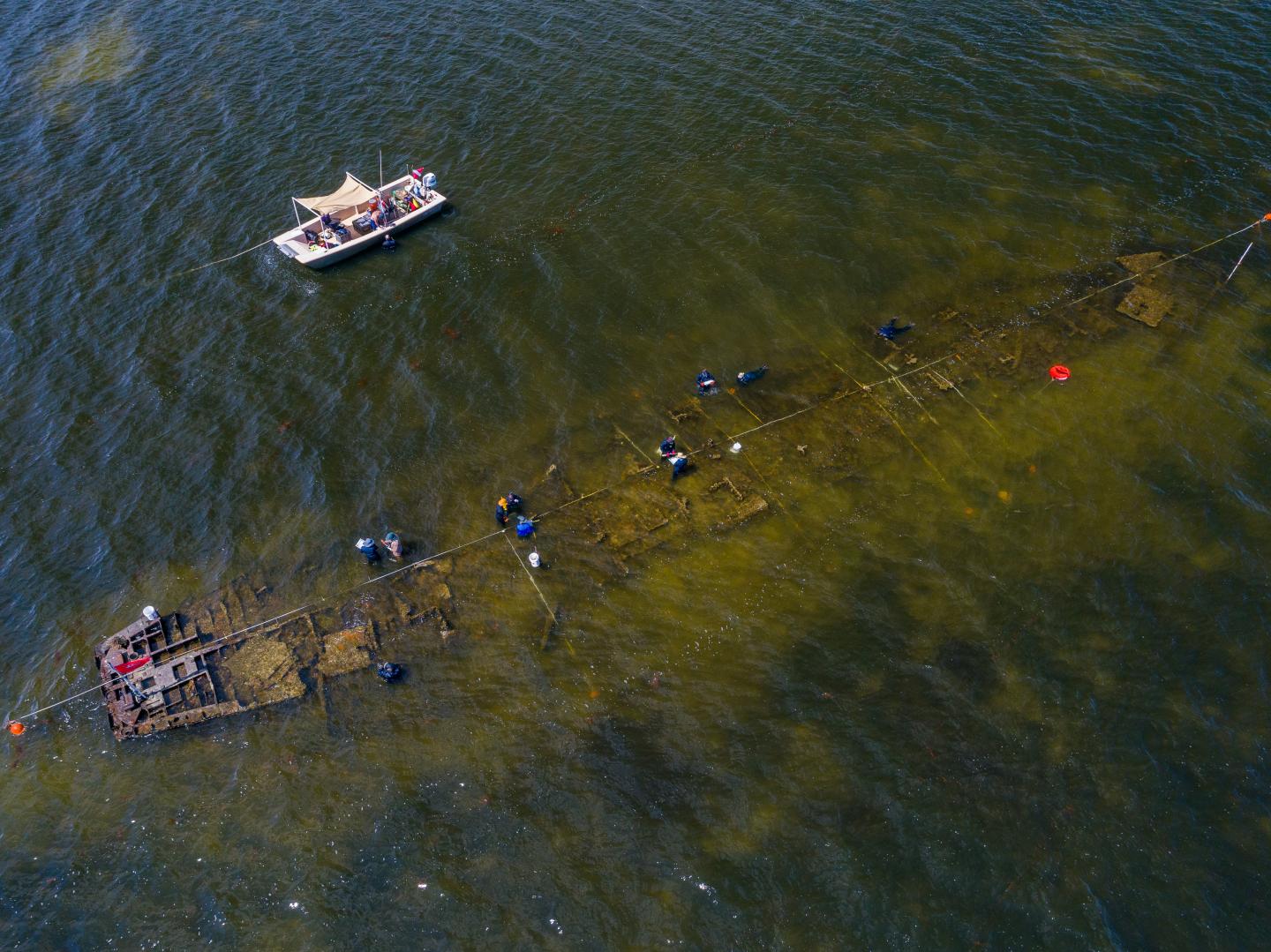
Credit: John McCord, Coastal Studies Institute
Shipwrecks act as artificial reefs and provide a substrate and nutrients for a great diversity of microorganisms, which can contribute to either the deterioration or preservation of the ship. Precisely how diverse such communities are, and how they are organized, is still unknown. Here, researchers from East Carolina University in Greenville, North Carolina, identify the bacteria associated with a shipwreck from the 1960s. They find a highly diverse community on the wreck, consisting of at least 4,800 OTUs (Operational Taxonomic Units, roughly corresponding to species) from 28 bacterial phyla, including nitrogen-, carbon-, sulfur-, and iron-cycling species. Microbial community composition strongly differed between locations within the site, suggesting niche partitioning, in the same way that fungal species specialize in particular microhabitats within a forest, based on the local abiotic and biotic environment. The results are published in the open-access journal Frontiers in Microbiology.
The 50-m-long wreck, called the Pappy Lane, represents the remains of the steel-hulled USS LCS(L)(3)-123, built in 1944 as a WWII warship and abandoned after running aground in the 1960s in the shallows of the Pamlico Sound lagoon, North Carolina, after a second career a barge. DNA sequencing of 14 samples from across the site – visibly corroded and visibly preserved shipwreck debris, drilled shipcores, nearby sediment, and surrounding seawater – revealed notable differences in composition and metabolic capacities of the local microbial communities living on and around the shipwreck, as well as the microbial communities living on different parts of the ship. The authors explain this diversity as evidence of niche partitioning, driven by small-scale variability in the abiotic environment, for example iron content, exposure to oxygen, and traces of hydrocarbons from a former fueltank.
Present across the shipwreck and abundant where corrosion was observed, were iron-oxidizing (“iron-eating”) Proteobacteria, which may contribute to biocorrosion. These included a new strain of the marine iron-oxidizing Zetaproteobacteria, with the apt name Mariprofundus ferrooxydans O1. Genomic analysis showed that the metabolic capacities of this strain include iron oxidation, carbon fixation in both oxygen-rich and -poor environments, and nitrogen fixation, indicating that it contributes to the cycling of metals and nutrients in the shipwreck environment.
This research also has wider implications for future resource management and the development of conservation strategies for shallow water shipwrecks across all coastlines.
“We have learned that iron-oxidizing bacteria that produce rust are widespread on these shipwrecks, causing corrosion and deterioration of the wreck-site. These microbes are more abundant in areas where we see corrosion occurring, which makes them likely indicators of where further deterioration may occur. In order to prevent this damage, we can design strategies for early detection, stopping their growth and limit further biocorrosion by other microbes,” says corresponding author Dr Erin Field, Assistant Professor in the Department of Biology at East Carolina University.
The results of this study point towards the need to adapt future conservation efforts to the unique situation of each shipwreck, taking into account original construction materials, environmental factors and time spent in water.
“Historically, shipwreck sites were treated as a single environment, but our research goes deeper, showing that there are different microbial communities within single wreck-sites and associated with the wreck itself. As such, we need to tailor conservation efforts to each shipwreck in order to more effectively mitigate biocorrosion and deterioration,” explains Dr Field.
This study highlights the importance of increasing the understanding of the role of biocorrosion in the deterioration of shipwrecks and the need for more research into the microbial ecosystem of shipwrecks.
“While there is well-developed literature on the impact of galvanic corrosion on shipwrecks and historic ships, the role certain microbes play in corrosion is less well-understood. It is hoped that this article helps decipher the mechanisms of biocorrosion that could one day also lead to the development of protective measures and conservation strategies,” concludes Dr Nathan Richards, Professor and Director of Maritime Studies in the Department of History at East Carolina University and co-author of the study.
###
Media Contact
Mischa Dijkstra
[email protected]
Related Journal Article
http://dx.





