New form of synthetic biology for controlling cellular behavior uses intrinsically disordered proteins
Credit: Michael Dzuricky, Duke University
DURHAM, N.C. – Biomedical engineers at Duke University have demonstrated a method for controlling the phase separation of an emerging class of proteins to create artificial membrane-less organelles within human cells. The advance, similar to controlling how vinegar forms droplets within oil, creates opportunities for engineering synthetic structures to modulate existing cell functions or create entirely new behaviors within cells.
The results appear online on August 3 in the journal Nature Chemistry.
Proteins function by folding into specific 3-D shapes that interact with different biomolecular structures. Researchers previously believed that proteins needed these fixed shapes to function. But in the last two decades, a large new class of intrinsically disordered proteins (IDPs) have been discovered that have large regions that are “floppy”–that is, they do not fold into a defined 3-D shape. It is now understood these regions play an important, previously unrecognized role in controlling various cellular functions.
IDPs are also useful for biomedical applications because they can undergo phase transitions–changing from a liquid to a gel, for example, or from a soluble to an insoluble state, and back again–in response to environmental triggers, like changes in temperature. These features also dictate their phase behavior in cellular environments and are controlled by adjusting characteristics of the IDPs such as their molecular weight or the sequence in which the amino acids are linked together.
“Although there are many natural IDPs that show phase behavior in cells, they come in many different flavors, and it has been difficult to discern the rules that govern this behavior,” said Ashutosh Chilkoti, the Alan L. Kaganov Distinguished Professor of Biomedical Engineering at Duke. “This paper provides very simple engineering principles to program this behavior within a cell.”
“Others in the field have taken a top-down approach where they’ll make a change to a natural IDP and see how its behavior changes within a cell,” said Michael Dzuricky, a research scientist working in the Chilkoti laboratory and first author of the study. “We’re taking the opposite approach and building our own artificial IDPs from simple thermodynamic principles. This enables us and others to precisely tune a single property–the shape of the IDPs phase diagram–to better understand how this parameter affects biological behavior”
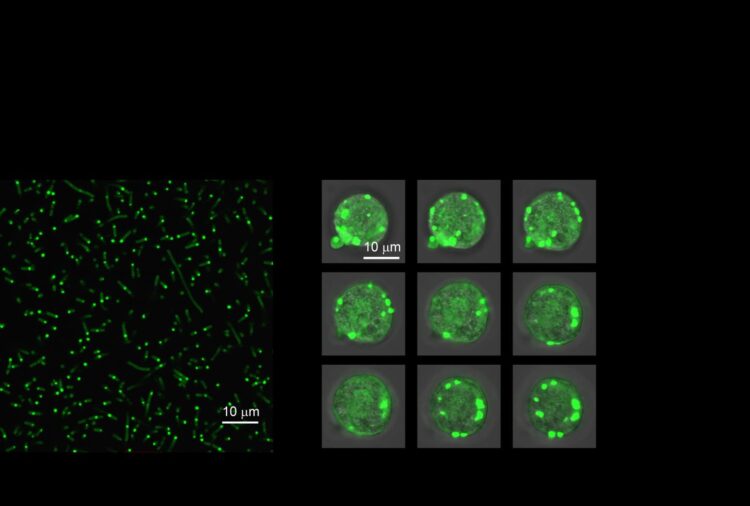
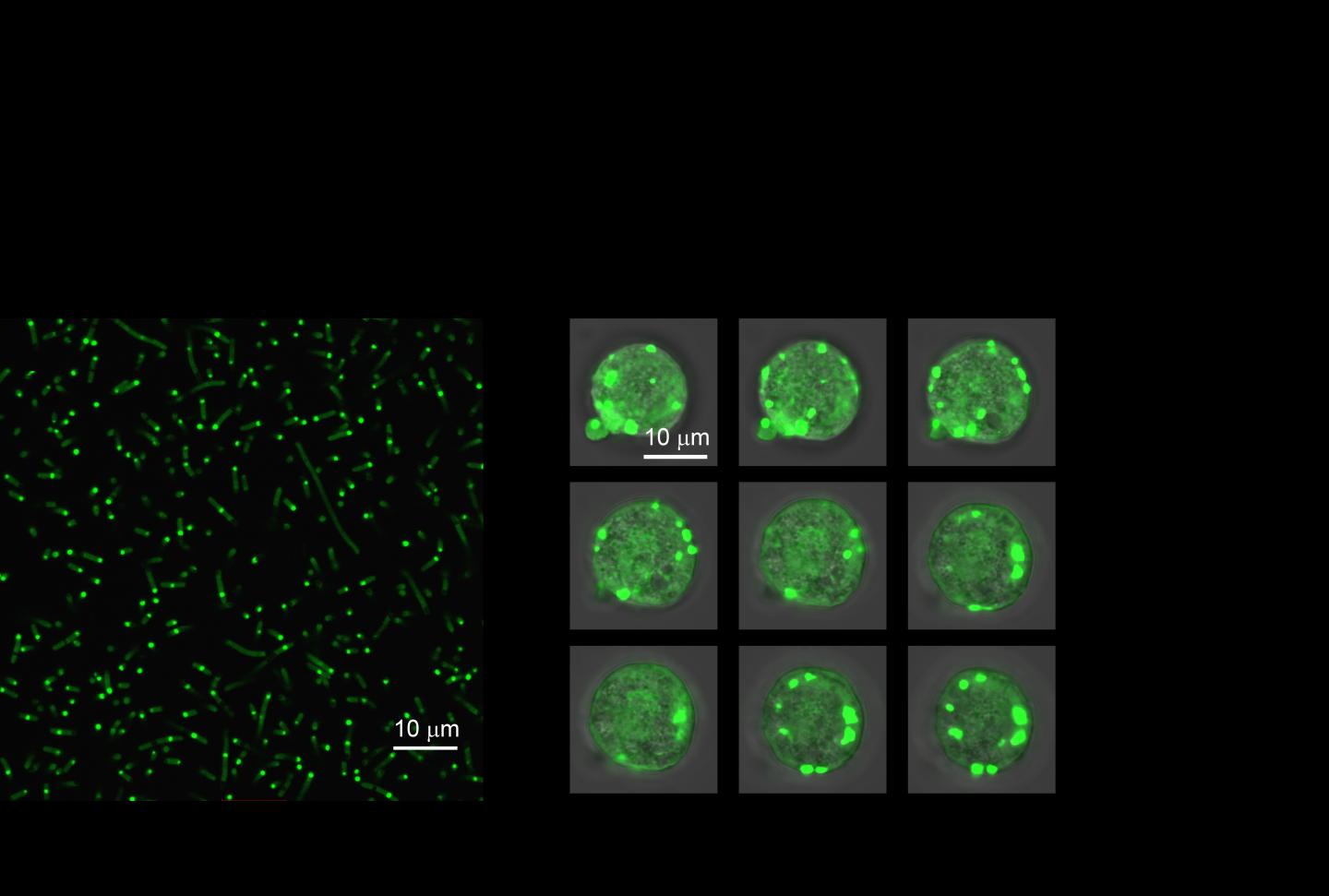
In the new paper, the researchers begin by looking to nature for examples of IDPs that come together to form “biomolecular condensates” within cells. These weakly-held-together structures allow cells to create compartments without also building a membrane to encapsulate it. Using one such IDP from the common fruit fly as a basis, the researchers draw from their extensive history of working with IDPs to engineer a molecularly simpler artificial version that retains the same behavior.
This simpler version allowed the researchers to make precise changes to the molecular weight of the IDP and amino acids of the IDPs. The researchers show that, depending on how these two variables are tweaked, the IDPs come together to form these compartments at different temperatures in a test tube. And by consistently trying various tweaks and temperatures, the researchers gained a solid understanding of which design parameters are most important to control the IDP’s behavior.
A test tube, however, is not the same as a living cell, so the researchers then went one step further to demonstrate how their engineered IDPs behave within E. coli. As predicted, their artificial IDPs grouped together to form a tiny droplet within the cell’s cytoplasm. And because the IDP’s behavior was now so well understood, the researchers showed they could predictably control how they coalesced using their test tube principles as a guide.
“We were able to change temperatures in cells to develop a complete description of their phase behavior, which mirrored our test tube predictions,” said Dzuricky. “At this point, we were able to design different artificial IDP systems where the droplets that are formed have different material properties.”
Put another way, because the researchers understood how to manipulate the size and composition of the IDPs to respond to temperature, they could program the IDPs to form droplets or compartments of varying densities within cells. To show how this ability might be useful to biomedical engineers, the researchers then used their newfound knowledge, as nature often does, to create an organelle that performs a specific function within a cell.
The researchers showed that they could use the IDPs to encapsulate an enzyme to control its activity level. By varying the molecular weight of the IDPs, the IDPs hold on the enzyme either increased or decreased, which in turn affected how much it could interact with the rest of the cell.
To demonstrate this ability, the researchers chose an enzyme used by E. coli to convert lactose into usable sugars. However, in this case, the researchers tracked this enzyme’s activity with a fluorescent reporter in real-time to determine how the engineered IDP organelle was affecting enzyme activity.
In the future, the researchers believe they could use their new IDP organelles to control the activity levels of biomolecules important to disease states. Or to learn how natural IDPs fill similar cellular roles and understand how and why they sometimes malfunction.
“This is the first time anybody has been able to precisely define how the protein sequence controls phase separation behavior inside cells,” said Dzuricky. “We used an artificial system, but we think that the same rules apply to natural IDPs and are excited to begin testing this theory.”
“We can also now start to program this type of phase behavior with any protein in a cell by fusing them to these artificial IDPs,” said Chilkoti. “We hope that these artificial IDPs will provide new tool for synthetic biology to control cell behavior.”
###
This research was supported by the National Institutes of Health (R35GM127042) and the National Science Foundation (DMR-17-29671, CHE-1709735).
“De Novo Engineering of Intracellular Condensates using Artificial Disordered Proteins.” Michael Dzuricky, Bradley A. Rogers, Abdulla Shahid, Paul S. Cremer, Ashutosh Chilkoti. Nature Chemistry, 2020. DOI: 10.1038/s41557-020-0511-7
Media Contact
Ken Kingery
[email protected]
Related Journal Article
http://dx.