Genome sequence data from three jellyfish species reveal evolutionary mechanisms of unique jellyfish traits and venomous toxins
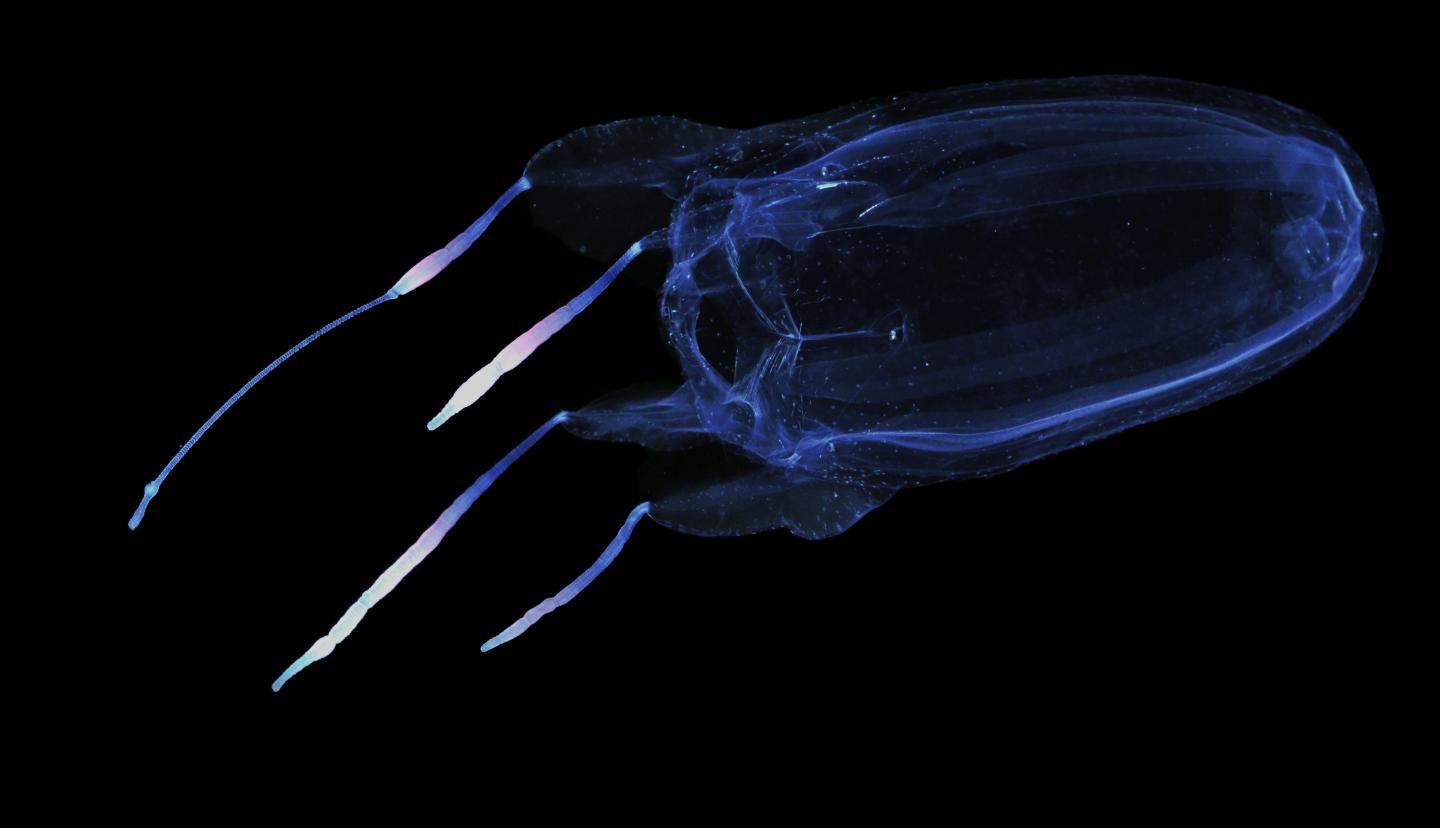
Credit: Allen Collins
An article published today in the Open Access journal GigaScience [1] might make you squirm if you plan to hit the beach this summer. The article presents the draft genomes of three different jellyfish species. The international group of researchers, lead by Joseph Ryan, chose to examine jellyfish that present a range of physical traits and level of toxicity (from minor annoyance to deadly) — jellyfish kill more people per year than the combined forces of sharks, stingrays, and sea snakes. Their research identified a range of venom related genes, providing the basis for exploring toxin gene evolution in these fascinating animals.
The three jellyfish species investigated are the deadly winged box jellyfish (Alatina alata), the anatomically interesting upside-down jellyfish (a true jellyfish: Cassiopea xamachana), and the stalked jellyfish (Calvadosia cruxmelitensis) so named for its stinging organs forming a Maltese Cross pattern. The work here finally provides genome sequences for all five lineages of the Medusozoa (a subphylum of Cnidaria).
Corresponding author Joseph Ryan said: “With these new genomes, for the first time, we were able to compare gene content of all 5 classes of cnidarians (Anthozoa, Hydrozoa, Cubozoa, Staurozoa, Scyphozoa). This broad survey provides an important overview of genomic evolution in cnidarians.”
Compared with advances in toxicological research in venomous vertebrates, toxicological research in cnidarians has been hampered by the lack of extensive genetic information. The area of cnidarian venom is of particular interest given that box jellyfish can be deadly to beach goers: approximately 100 people die each year from their stings. They are considered so dangerous that they are the only jellyfish to have its own category in the US Weather Report.
Ryan notes: “Box jellyfish are among the most venomous animals on the planet and therefore their genomes are important resources for developing antivenoms as well as potential drugs.” He added, “Our survey included a focused assessment of venom-related genes, providing insight into the evolution of cnidarian venom.”
To investigate the toxins, the authors identified 117 putative venom proteins, of which 9 were found only in cnidarians. The winged box jellyfish genome was the only one to include a toxin gene called CqTx. The CqTx protein creates pores in the membranes of cells resulting in hemolysis, the breaking down of red blood cells. Given that some box jellyfish can stop someone’s heart within 5 minutes, the identification of this toxin in the winged box jellyfish adds another clue to the still unclear mechanisms of how people die from a box jellyfish sting.
Comparisons of the genomes also revealed information about the evolution of cnidarian venom. Gene loss and gene duplication both play large roles in species evolution, and, as an example, the scientists’ analysis of another toxin gene, CrTx, showed both. The upside-down jellyfish contained a single CrTx gene, the winged box jellyfish had five copies of this gene; and the stalked jellyfish had none.
Ryan expands on these findings, saying: “Gene loss is an important driver of evolutionary change. Having draft genomes allows for accurate inferences of gene loss and for understanding the phenotypic contribution of gene loss in these animals. Furthermore, these genomes provide information about the order of genes in the genome, which in some cases allow for independent verification of the identification of hard-to-classify genes.”
The addition of these three new jellyfish genome sequences means that there are now genome sequences available in all five classes of cnidarians. Having access to this wealth of genetic information will enable the exploration of the genetic underpinnings of this group’s incredibly diverse range of biological traits, which includes, but isn’t limited to, differences in life cycles, prey preferences, metamorphoses, swimming behaviors, population blooms, sexual behaviors, environmental stress reactions, cell type determination, and stinging capsule development. Truly, within Cnidaria, researchers have a — now unburied — treasure chest of information.
###
Further Reading:
1. Aki Ohdera, Cheryl L. Ames, Rebecca B. Dikow, Ehsan Kayal, Marta Chiodin, Ben Busby, Sean La, Stacy Pirro, Allen G. Collins, Mónica Medina, Joseph F. Ryan. Box, stalked and upside-down? Draft genomes from diverse jellyfish (Cnidaria, Acraspeda) lineages: Alatina alata (Cubozoa), Calvadosia cruxmelitensis (Staurozoa), and Cassiopea xamachana (Scyphozoa). GigaScience, 2019. https:/
Publication in GigaScience includes storage of relevant associated data in the journal’s affiliated database, GigaDB, where every dataset is provided with a digital object identifier (DOI), making it possible to cite and track data in standard scientific literature, which serves as a strong incentive for researchers to more rapidly release expensive and work-intensive datasets for community use.
The data citation for this article is:
Ohdera AH; Ames CL; Dikow RB; Kayal E; Chiodin M; Busby B; La S; Pirro S; Collins AG; Medina M; Ryan JF: Supporting data for “Boxed, stalked and upside-down? Draft genomes from diverse jellyfish (Cnidaria, Acraspeda) lineages: Alatina alata (Cubozoa), Calvadosia cruxmelitensis (Staurozoa), and Cassiopea xamachana (Scyphozoa)” GigaScience Database. 2019. http://dx.
GigaScience is co-published by BGI, the world’s largest genomics organization, and Oxford University Press. The journal covers research that uses or produces ‘big data’ from the full spectrum of the life sciences. It also serves as a forum for discussing the difficulties of and unique needs for handling large-scale data from all areas of the life sciences. The journal has a completely novel publication format — one that integrates manuscript publication with complete data hosting, and analyses tool incorporation. To encourage transparent reporting of scientific research as well as enable future access and analyses, it is a requirement of manuscript submission to GigaScience that all supporting data and source code be made available in the GigaScience database, GigaDB , as well as in their publicly available repositories. GigaScience will provide users access to associated online tools and workflows, and has integrated a data analysis platform, maximizing the potential utility and re-use of data.
Follow GigaScience on twitter @GigaScience; Facebook https:/
Media Contacts
Nicole Nogoy
Editor, GigaScience, BGI Hong Kong
Tel: +852 3610 3531
Mob: +64 22 698 2123
Email: [email protected]
Media Contact
Nicole Nogoy
[email protected]
Related Journal Article
http://dx.




