IBS-developed Digenome-seq technique reveals the accuracy of CRISPR-based editing tool that changes DNA letter A to G
Credit: IBS
Researchers at the Center for Genome Engineering, within the Institute for Basic Science (IBS, South Korea) have identified the mistake-rate of DNA editing tools, based on CRISPR and known as adenine base editors. Assessing the genome-wide target specificity of these innovative techniques is essential to harness their applications in clinics and biotechnology. Their findings were published in Nature Biotechnology.
DNA’s four letters, or bases, are the alphabet used by our cells: adenines (A) pairs with thymines (T), cytosines (C) with guanines (G), making a unique combination of 3.2 billion letters, that makes us who we are. Since some genetic diseases are caused by a mutation of just one letter, some of the applications of CRISPR – a very successful and powerful gene engineering tool – deal with the correction of this single-letter difference. Examples of proteins that can be added to the CRISPR system to promote letter conversions are: cytosine base editors (CBEs) for C-to-T conversions, and adenine base editors (ABEs) for A-to-G changes. The IBS team has been interested in studying ABEs’ specificity, as it has not been known so far.
The team, led by Jin-Soo Kim, studied the error-rate of recently developed ABE proteins, ABE7.10, in human cells. They pinpointed the positions on the human genome affected by ABE7.10 and scanned for errors beyond the target. To do that, they used an adapted version of Digenome-seq, a sequencing technique developed by the same Research Center, that had already successfully determined the accuracy of CBE, CRISPR/Cas9 and CRISPR/Cpf1, among others. They tested ABE7.10 with seven guide RNAs, corresponding to seven DNA target letters, and also compared the results with a common CBE, and a Cas9 nuclease. The modified Digenome-seq could detect an average of 60 off-target mistakes in the entire human genome. And interestingly, although the three proteins were engineered to target the same site, they recognized different off-target points.
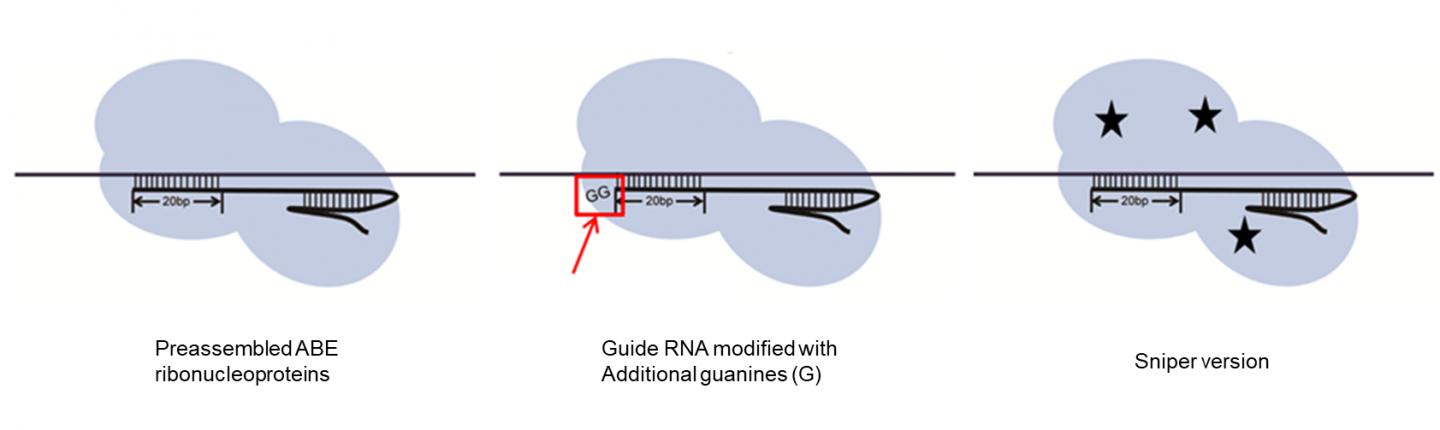
IBS biologists also showed some strategies to curb the number of off-target modifications. Adding a couple of Gs at the end of the guide RNA reduced the off-target mistakes, as well as the use of a different type of Cas9 (Sniper-Cas9, developed by the same team in 2018) and the delivery of ABE7.10 via preassembled ribonucleoproteins, rather than via plasmids.
The team aims to contribute to the development of ABEs, to introduce the desired single-letter changes in a more precise and efficient way. “As the accuracy of the base editor is proven, we expect that it will find wide application in the future in medical and agricultural realms,” says Jin-Soo Kim.
###
Media Contact
Dahee Carol Kim
[email protected]
Related Journal Article
http://dx.




