Credit: Almary Guerra
Scientists have shown for the first time that the blueprint for the four chambers of the human heart exists in the humble zebrafish, a study in eLife reports.
The discovery provides key insights into processes that go wrong in congenital heart defects and means that researchers now have a simple model for further research that more closely reflects what happens in human heart development.
During development, the heart forms four chambers that support blood flow to the lungs and the rest of the body. The arrangement of these parallel blood systems was essential for the evolution of mammals from aquatic ancestors, but the processes by which the left and right cardiac compartments are separated remain poorly understood.
Research has shown that formation of the adult heart occurs in part from two sets of precursor cells called the first heart field (FHF) and the second heart field (SHF). FHF cells form most of the left ventricle, whereas cells from the SHFs are involved in shaping the right ventricle. By contrast, both left and right atria involve cells from both groups.
"This suggested that the left and right sides of the ventricle and atrium of the heart are developed by different biological processes, to ensure distinction between the side that supports the lungs, and the side that supports blood flow to the rest of the body," explained Dr Almary Guerra, graduate student at the Max Planck Institute for Heart and Lung Research, Germany, and lead author of the study. "But given that the same cells are involved in both sides of the atrium, we still do not understand how the left and right parts of the atrium develop separately from each other."
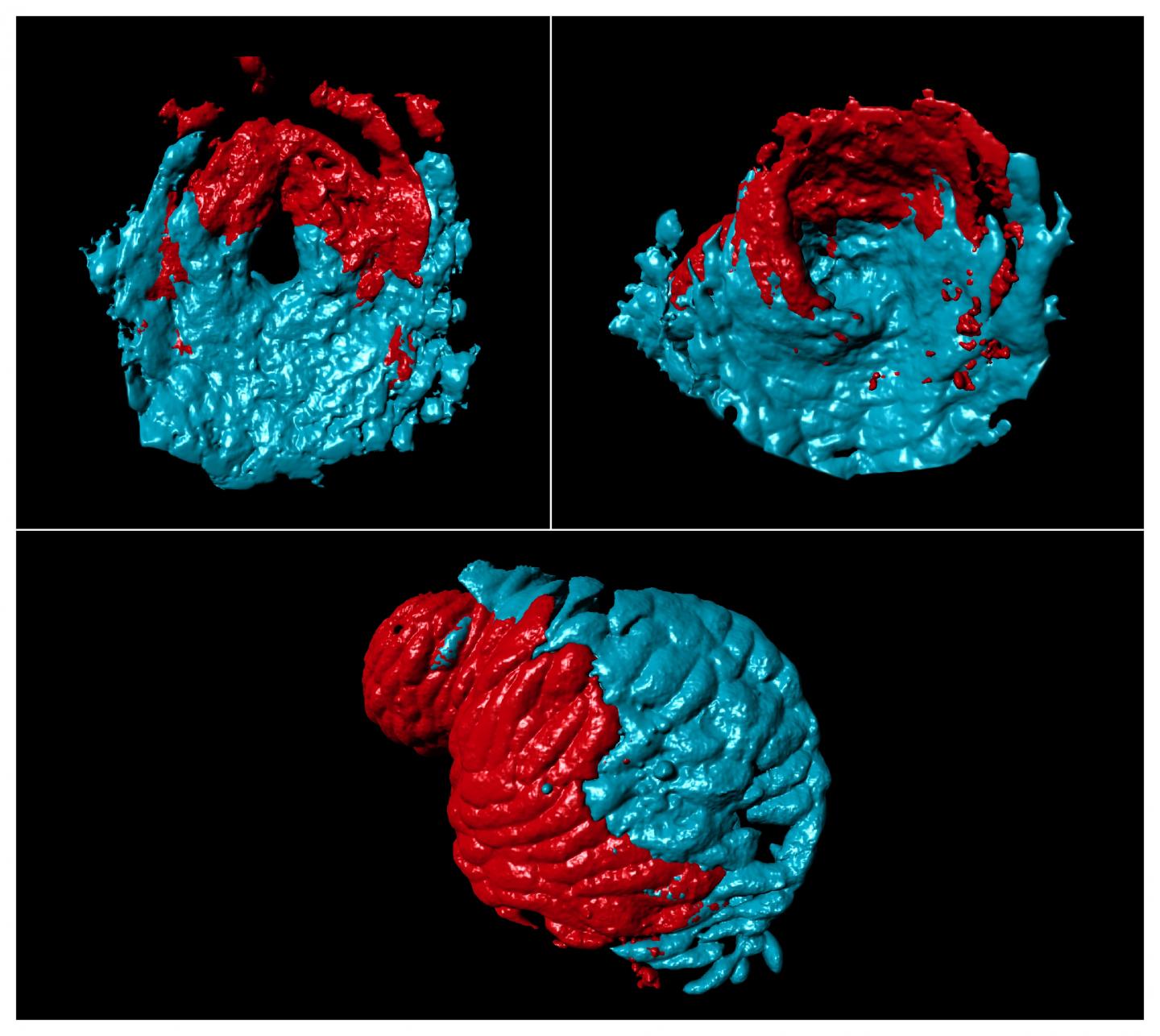
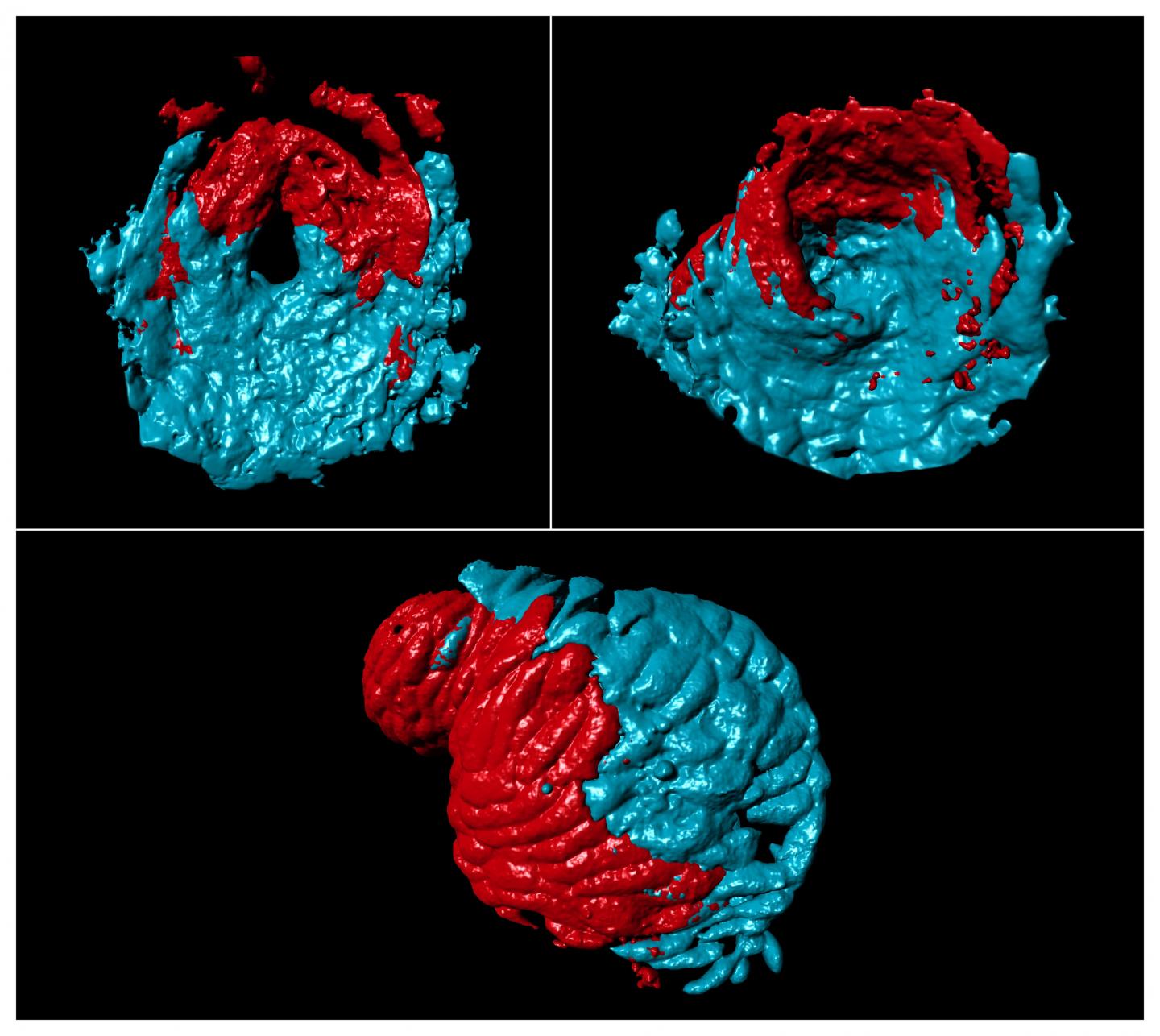
Studies of zebrafish have contributed significantly to our understanding of heart development, and Dr Guerra and colleagues used this organism to explore whether there were differences in molecules and cells between the two sides of the single atrium.
They first looked at which genes were most active in the atrium and found two of interest called pitx2c and meis2b. Mutations in the human versions of both genes are known to cause defects in the formation of the heart septum, a serious condition which demands surgical intervention. meis2b was found in different amounts in the left and right sides of the heart, and both pitx2c and meis2b were most often found in the same place. Further analysis revealed that meis2b controls pitx2c expression, revealing an important insight for potential treatment of heart defects.
To work out the origin of this gene pattern, the team studied genes in different regions of the heart as the zebrafish developed over time. They found that meis2b was most active in a group of cells that form the left side of the heart during the earliest stage of development and this was maintained through to adulthood. Moreover, the SHF precursor cells involved in atrial formation were present in regions where meis2b was both present and absent, confirming that SHF contributes equally to both sides of the atrium and is not involved in asymmetric development of the atria.
Finally, to understand whether meis2b was essential for correct atrial function, they mutated the gene in zebrafish and studied its effects. In early development, there was an overgrowth of the atrium in relation to the ventricle, which was due to an increase in heart-cell division. ECG tests on the zebrafish revealed that the mutation adversely affected the electrical activity of the heart, similarly to what is known in humans.
"We have shown for the first time that atrial compartmentalization occurs in the zebrafish," said senior author Dr Sven Reischauer, Junior Group Leader at the Max Planck Institute for Heart and Lung Research. "The next step is to identify the specific elements that control atrial asymmetry and to use the model to reveal further insights into heart development that will be invaluable for understanding and treating heart defects."
###
Reference
The paper 'Distinct myocardial lineages break atrial symmetry during cardiogenesis in zebrafish' can be freely accessed online at https://doi.org/10.7554/eLife.32833. Contents, including text, figures and data, are free to reuse under a CC BY 4.0 license.
Media contact
Emily Packer, Senior Press Officer
eLife
[email protected]
01223 855373
About eLife
eLife aims to help scientists accelerate discovery by operating a platform for research communication that encourages and recognises the most responsible behaviours in science. We publish important research in all areas of the life and biomedical sciences, which is selected and evaluated by working scientists and made freely available online without delay. eLife also invests in innovation through open-source tool development to accelerate research communication and discovery. Our work is guided by the communities we serve. eLife is supported by the Howard Hughes Medical Institute, the Max Planck Society, the Wellcome Trust and the Knut and Alice Wallenberg Foundation. Learn more at https://elifesciences.org.
Media Contact
Emily Packer
[email protected]
@elife
http://www.elifesciences.org
Related Journal Article
http://dx.doi.org/10.7554/eLife.32833