Credit: Chan Cao, EPFL
Some types of bacteria have the ability to punch holes into other cells and kill them. They do this by releasing specialized proteins called “pore-forming toxins” (PFTs) that latch onto the cell’s membrane and form a tube-like channel that goes through it. This hole (structure?) across the membrane is called a pore. Punctured by multiple PFTs, the target cell self-destructs.
However, PFTs have garnered much interest beyond bacterial infections. The nano-sized pores that they form are used for “sensing” biomolecules: a biological molecule e.g. DNA or RNA, passes through the nanopore like a string steered by a voltage, and its individual components (e.g. nucleic acids in DNA) give out distinct electrical signals that can be read out. In fact, nanopore sensing is already on the market as a major tool for DNA or RNA sequencing.
Publishing in Nature Communications, scientists led by Matteo Dal Peraro at EPFL have studied another major PFT that can be used effectively for more complex sensing, such as protein sequencing. The toxin is aerolysin, which is produced by the bacterium Aeromonas hydrophila, and is the “founding member” of a major family of PFTs found across many organisms.
One of the main advantages of aerolysin is that it forms very narrow pores that can tell apart molecules with much higher resolution than other toxins. Previous studies have shown that aerolysin can be used to “sense” several biomolecules, but there haven’t been barely any studies on the relationship between aerolysin’s structure and its molecular sensing abilities.
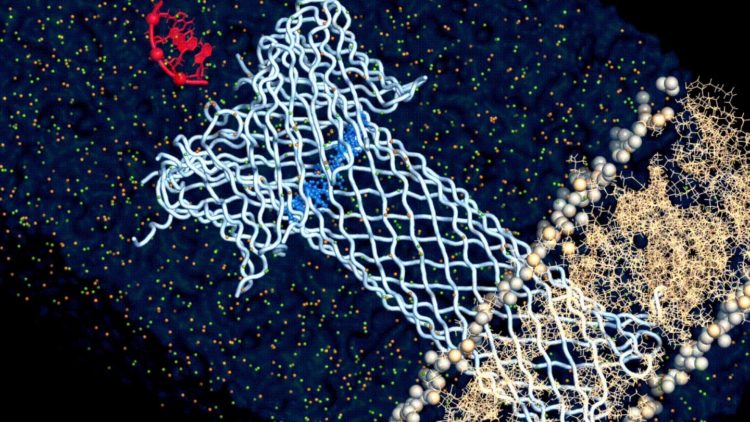
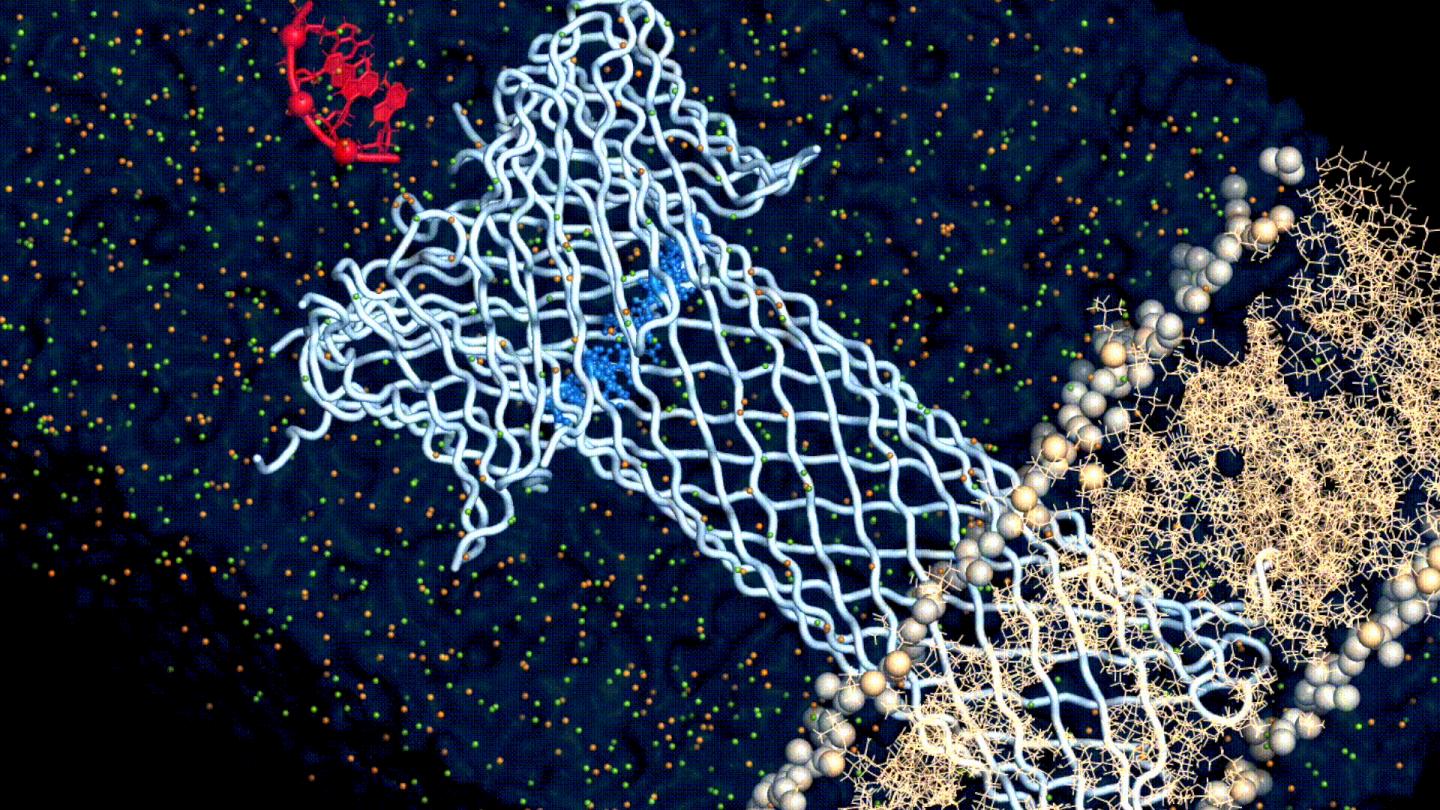
The researchers first used a structural model of aerolysin to study its structure with computer simulations. As a protein, aerolysin is made up of amino acids, and the model helped the scientists understand how those amino acids affect the function of aerolysin in general.
Once they had a grasp of that relationship, the researchers began to strategically change different amino acids in the computer model. The model then predicted the possible impact of each change on the overall function of aerolysin.
At the end of the computational process, Dr Chan Cao, the leading author of this work, produced sixteen genetically engineered, “mutant” aerolysin pores, embedded them in lipid bilayers to simulate their position in a cell membrane, and carried out various measurements (single-channel recording and molecular translocation experiments) to understand how ionic conductance, ion selectivity, and translocation properties of the aerolysin pore are regulated on a molecular level.
And with this approach, the researchers finally found what drives the relationship between the structure and the function of aerolysin: its cap. The aerolysin pore isn’t just a tube that goes through the membrane, but also has a cap-like structure that attracts and tethers the target molecule and “pulls” it through the pore’s channel. And the study found that the it is the electrostatics at this cap region that dictate this relationship.
“By understanding the details of how the structure of the aerolysin pore connects to its function, we can now engineer custom pores for various sensing applications,” says Dal Peraro. “These would open new, unexplored opportunities to sequence biomolecules as DNA, proteins and their post-translational modifications with promising applications in gene sequencing and biomarkers detection for diagnostics.” The scientists have already filed a patent for their sequencing and characterization of the genetically engineered aerolysin pores.
###
Other contributors
Federal University of Rio de Janeiro
Skolkovo Institute of Science and Technology (Russia)
EPFL Laboratory of Nanoscale Biology
Reference
Chan Cao, Nuria Cirauqui, Maria Jose Marcaida, Buglakova Elena, Alice Duperrex, Aleksandra Radenovic, Matteo Dal Peraro. Single-molecule sensing of peptides and nucleic acids by engineered aerolysin nanopores. Nature Communications 29 October 2019. DOI: 10.1038/s41467-019-12690-9
Media Contact
Nik Papageorgiou
[email protected]
41-216-932-105
Related Journal Article
http://dx.