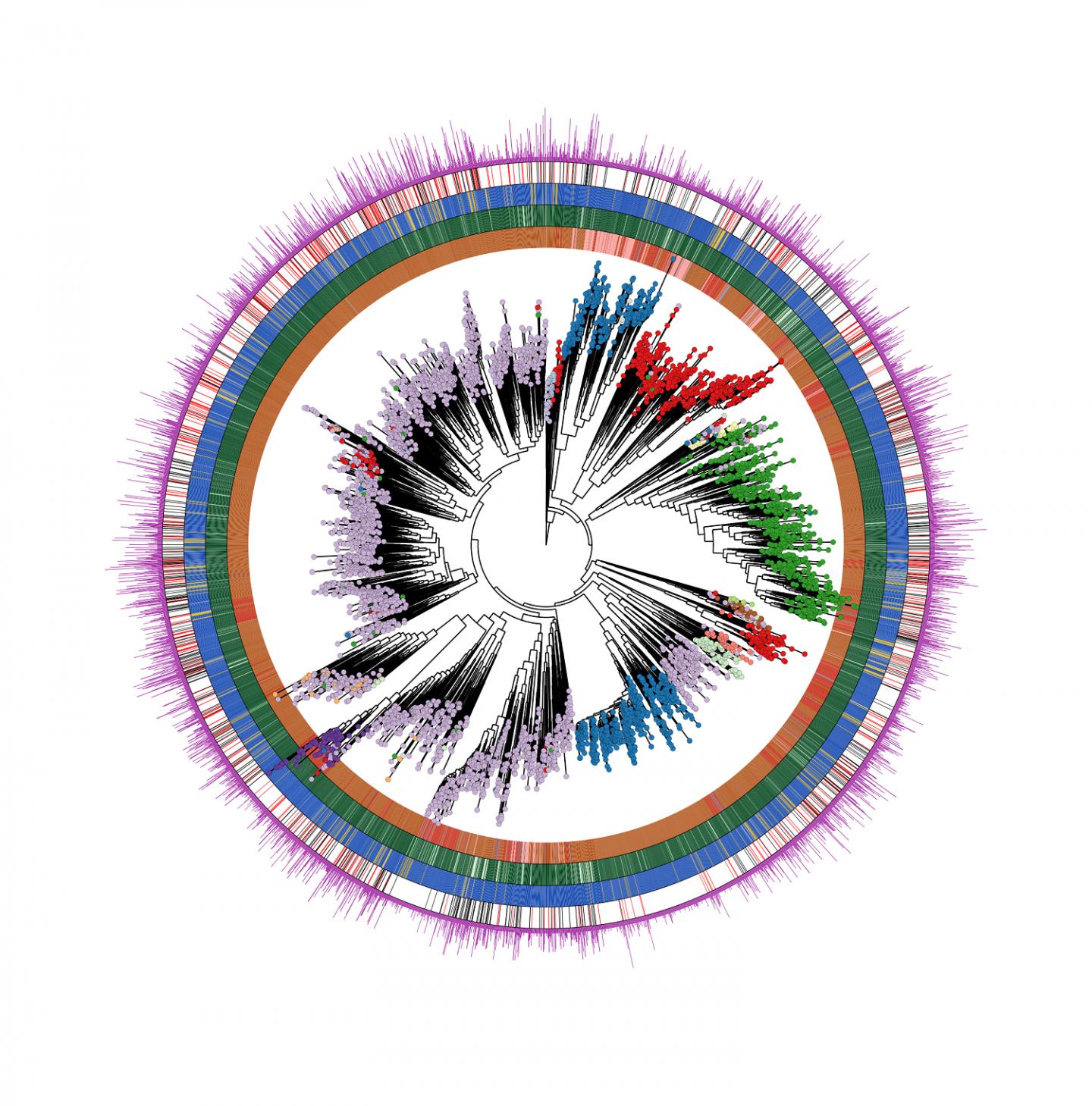
A large number of previously uncharacterized bacteria living in and on humans was discovered and analyzed by scientists at the University of Trento, Italy. This computational metagenomic study appeared in “Cell”
Credit: ©University of Trento
Dr. Segata explains: «We genetically characterized and catalogued a large number of bacteria and archaea that are part of the human microbiome, but remained so far unexplored, uncharacterized, and undescribed. We also observed that many of these microbes tend to be only rarely identified in Westernized populations, most probably as a indirect consequence of the complex industrialization processes».
Similarly to other living organisms, microbes evolve and are under selection pressure as the environment, including diet and lifestyle, changes. In some cases, human-associated microbes that can be crucial for our health are at risk of extinction. The research team investigated some of these cases. The presented study integrates genomics, microbiology, and big data and engaged several researchers at the CIBIO Department of the University of Trento as well as students of the masters program in Quantitative and Computational Biology at the same institute. The study also involved partners at Harvard University (US) that were responsible for the microbiome sampling of a Malagasy population, and at Otago University (New Zealand), National Research Council in Spain (Valencia), and University of Warwick (UK).
Nicola Segata focuses his research on the human microbiome. He explains: «It’s the collection of bacteria, archaea, viruses, fungi, and parasites that populate human body sites such as the gut, the mouth, the skin, and the urogenital tract. The human microbiome is in symbiosis with our own cells and plays a key role for our health, for example, in the metabolism of dietary compounds, in regulating the so-called gut-brain-axis, in protecting us from pathogenic agents, and in modulating our immune system. It has also been recently shown that the microbiome is involved in the etiology of some cancers and in the success of anti-tumoral immunotherapy approaches».
The approach of his team in studying the human microbiome is called “computational metagenomics”: they study the microbiome by analyzing its genetic information. From a drop of saliva, a skin swab, or a gram of stool they extract the total DNA of the microbes in the sample, and they subject the DNA to high-throughput sequencing. The resulting huge amount of genetic data is analyzed with specialized software to reconstruct the genomes of the microbes present in the microbiome.
Dr. Segata details some aspects of the new study: «Our findings are the result of a multidisciplinary team at CIBIO comprising microbiologists, statisticians, and computer scientists that identified a total of almost 5,000 microbial species recapitulating over 154,000 newly reconstructed genomes describing the human microbiome across ages, body sites, lifestyles, and diseases. Each of us is colonized by several hundreds of these species. But a large fraction of them (77%) were previously unknown. Many of these species are relatively rare, but some are very prevalent in human populations worldwide, and their discovery is the starting point for testing their potential role in autoimmune, gastrointestinal, and oncologic diseases. To obtain these results, we analyzed an extremely large dataset of publicly available and newly obtained microbiome samples spanning geography, population lifestyles, and age. Overall, we considered 9,428 human microbiome samples that have been studied using DNA sequencing technology called metagenomics».
A collaborator on the study, Dr. Curtis Huttenhower of the Harvard T.H. Chan School of Public Health, commented that, “Very large-scale studies of the human microbiome have begun to be particularly important for understanding population health. A surprising finding about the microbiome has been how personalized it is, though, and the level of detail necessary to understand its influence on each individual’s health. Techniques such as those developed in this study have the ability to identify new microbes unique to each person, and the genes and molecules unique to those microbes, that can in turn carry out immune signaling or modify dietary processes in the gut. The fact that we found such striking differences around the world during this work also calls out the microbiome’s relevance to global health – for example, in explaining some of the differences in response to vaccines that might be attributable to microbial rather than human genetic diversity.”
On the discovery and analysis of one of these bacteria, Segata continues: «The most common previously unknown candidate species, that we called “Cibiobacter qucibialis”, is the seventh most prevalent human-associated microbe in the worldwide population. We studied it by reconstructing more than 1,800 genomes. We think this species could be of particular relevance for further understanding the functions of the human microbiome».
And a final highlight on the observed differences among human populations: «We specifically focused on non-Westernized populations that do not have access to high-fat diet, common drugs including antibiotics, and do not live in highly sanitized environments. Many novel microbes discovered in non-Westernized populations across continents tend to be almost undetectable in Westernized populations. Our work thus enabled the study of such microbial species that could potentially be linked in the future with the increasing incidence of autoimmune diseases, allergies, and complex syndromes in the Westernized populations. It will thus be crucial to isolate, cultivate, and maintain these species that could be hypothetically reintroduced in Westernized populations with novel intervention strategies».
###
Media Contact
SALETTI ALESSANDRA
[email protected]
Original Source
https:/
Related Journal Article
http://dx.