A team of scientists, including several from the U.S. Department of Energy's Argonne National Laboratory, have determined the structures of several important tuberculosis enzymes, which could lead to new drugs for the disease.
Tuberculosis, caused by Mycobacterium tuberculosis bacteria, has proved incredibly stubborn even in the age of powerful antibiotics, infecting about one third of all people worldwide. Treatment can take up to nine months. It has stealth properties that protect it from antibiotics; it can hide inside human cells, avoiding the body's immune system while it waits for the opportune moment to multiply; and it's very resourceful at acquiring resistance.
"What we have now may not work in a few years," said Andrzej Joachimiak, an Argonne Distinguished Fellow, head of the Structural Biology Center, co-principal investigator at the Center for Structural Genomics of Infectious Diseases and a corresponding author on the new study.
In order to make new drugs, researchers need to search through the thousands of proteins in the bacterial world to find one that does something so important the bacterium can't live without it–and then make a little block to match.
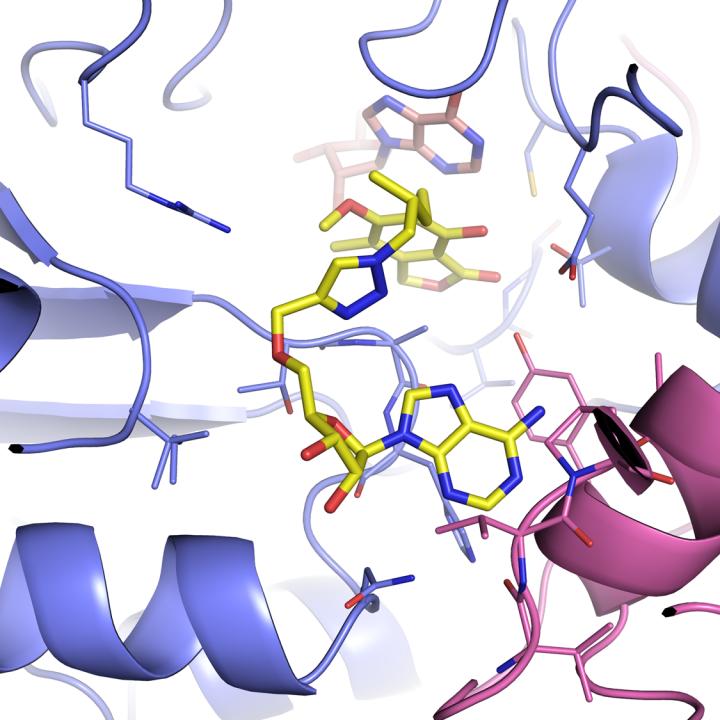
One such entry point might be IMPDH (inosine-5?-monophosphate dehydrogenase), which is part of a cellular process that controls the making of guanine nucleotides, one of the building blocks for DNA and RNA. It's so essential that virtually all living organisms, including human and bacterial pathogens, have versions of it.
"What we discovered earlier this year is that the human and bacterial versions bind molecules differently," Joachimiak said. "This is very important for finding a molecule to build a drug around–you don't want to inhibit a human enzyme, just the pathogen one."
Researchers have been interested in the mycobacterium IMPDH enzyme as a drug target for years, Joachimiak said, but haven't been able to produce it well enough to study it.
The team observed that one part of the enzyme's structure was particularly wobbly, so they engineered a version without it using resources at the Advanced Protein Characterization Facility and then then determined the structure employing synchrotron protein crystallography at the Advanced Photon Source, a DOE Office of Science User Facility (both at Argonne).
The modified version functions very similarly to the original, Joachimiak said, but is much easier to purify and crystallize for study.
Brandeis University professor Lizbeth Hedstrom and University of Minnesota professor Courtney Aldrich, two of the study's other research collaborators, had identified several inhibitor molecules that bind to IMPDH, and thus might be a starting point for a drug–but they couldn't be imaged while interacting with the enzyme. The new engineered enzyme allowed them to capture the structures of Hedstrom's and Aldrich's inhibitors in action, locked with IMPDH.
Helena Boshoff at the National Institute of Allergies and Infectious Diseases performed complementary studies showing that these inhibitors do in fact efficiently block mycobacterium growth.
The new structures were deposited into the Protein Data Bank for continued study.
###
The study, "Mycobacterium tuberculosis IMPDH in Complexes with Substrates, Products and Antitubercular Compounds," was led by Magdalena Makowska-Grzyska with the University of Chicago's Center for Structural Genomics of Infectious Diseases (CSGID); Argonne and CSGID protein crystallographer Youngchang Kim also co-authored the study. Other collaborators included researchers with Brandeis University, the National Institute of Allergies and Infectious Diseases (NIAID), the University of Minnesota, and the University of Houston. The research was supported by the National Institutes of Health and NIAID. Use of the Structural Biology Center was supported by the DOE Office of Science.
Argonne National Laboratory seeks solutions to pressing national problems in science and technology. The nation's first national laboratory, Argonne conducts leading-edge basic and applied scientific research in virtually every scientific discipline. Argonne researchers work closely with researchers from hundreds of companies, universities, and federal, state and municipal agencies to help them solve their specific problems, advance America's scientific leadership and prepare the nation for a better future. With employees from more than 60 nations, Argonne is managed by UChicago Argonne, LLC for the U.S. Department of Energy's Office of Science.
The U.S. Department of Energy's Office of Science is the single largest supporter of basic research in the physical sciences in the United States and is working to address some of the most pressing challenges of our time. For more information, visit the Office of Science website.