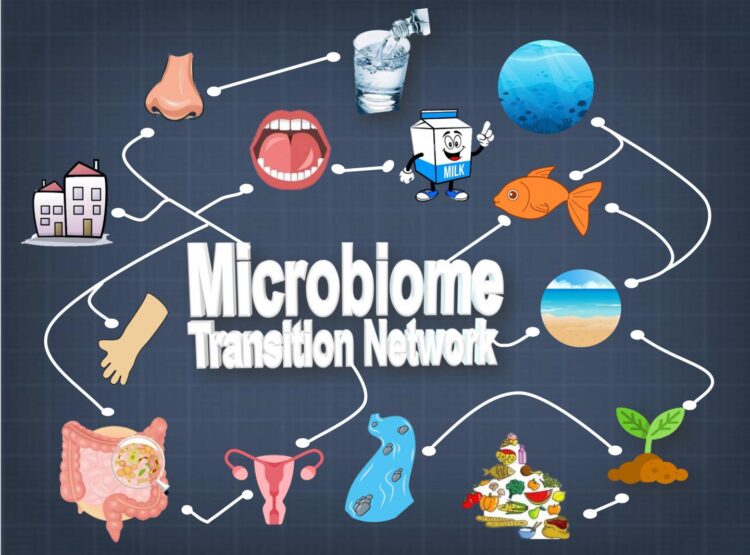
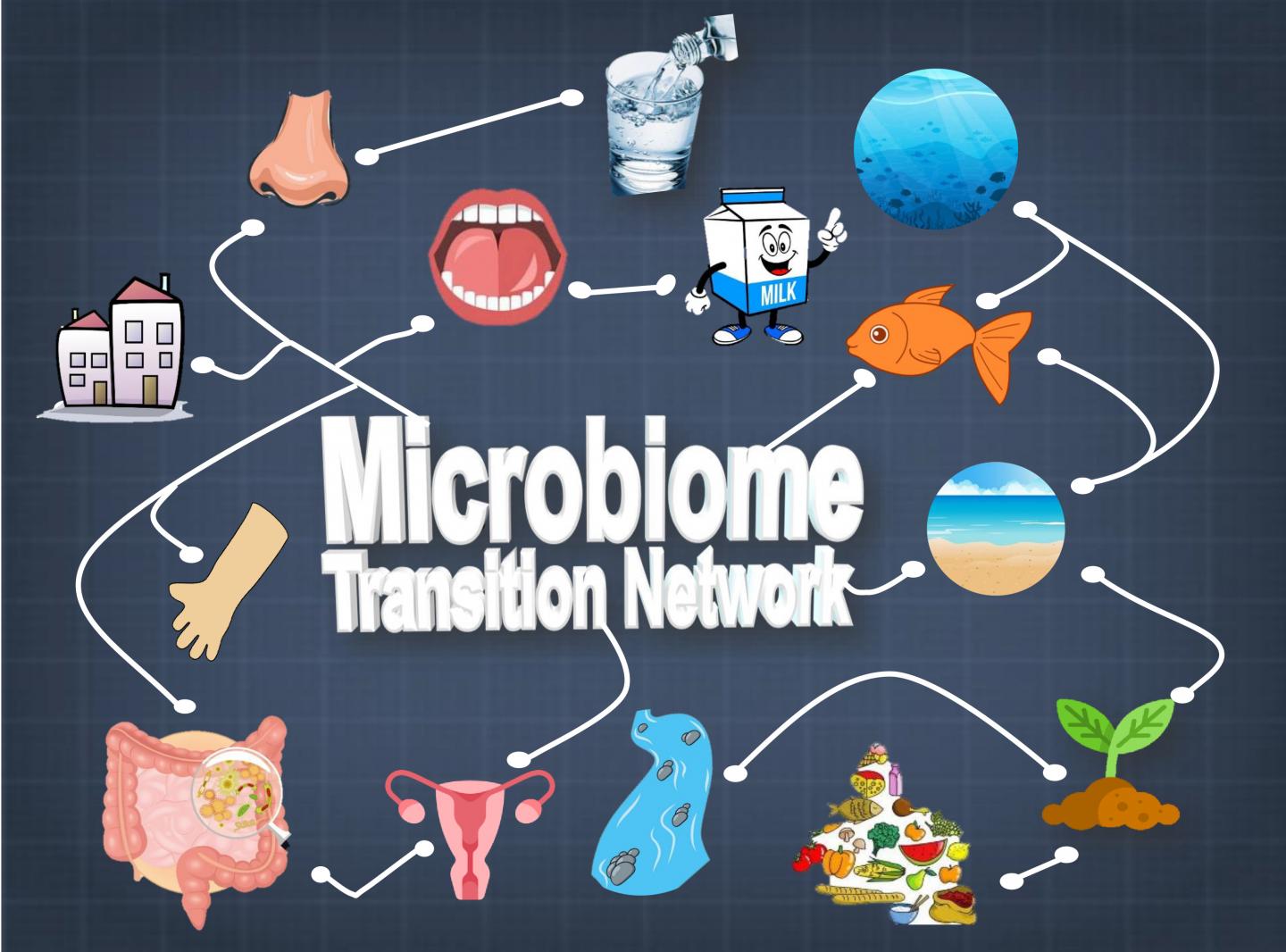
Global Microbiome Transition Network hints at origins and evolution paths of microbiomes
Credit: LIU Yang and JING Gongchao
The Earth contains about one trillion species of microbes — only about one-tenth of which have been identified. A single human can house 100 trillion microbes, creating a single microbiome that serves an ecosystem of microbes.
Microbes connect and transform in myriad ways, creating and combining and separating microbiomes anew. How can we begin to parse out how microbiomes differ, how they are similar, how they evolved and how they may change in the future?
An international team of researchers may have the answer. They published a scale-free, fully connected search-based network to explore the connectedness of microbiomes across the world on July 13 in mSystems.
“The microbiome composition, a fundamental feature of all microbiota — microbes sharing a particular characteristic such as site or geological period — is shaped by a plethora of environmental factors,” said co-first author JING Gongchao, a researcher in the Single-Cell Center in Qingdao Institute of BioEnergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences (CAS). “However, it remains unclear whether and how compositional changes at the ‘community-to-community’ level among microbiomes are linked to the origin and evolution of global microbiome diversity.”
To better understand how the vast number of varying microbe species evolved, JING and ZHANG Yufeng, a graduate student from Qingdao University, built a global “microbiome transition network” (MTN) that connects, based on their composition similarities, 177,022 microbiomes from 20 diverse ecosystems that include the plethora of ecological niches on human body and in the environment.
They used the Microbiome Search Engine (MSE; http://mse.
Interestingly, the global MTN is scale-free, which is similar to the Internet or the social network among human individuals. In this kind of networks, most nodes are each connected to a small number of other nodes, yet a small portion of nodes are connected to many other nodes. Such network structure ensures a strong degree of tolerance of the network against accidental perturbation.
“We drew the first global microbiome transition roadmap to illustrate the potential yet most likely paths to explain the evolution process of global microbiomes,” explained SU Xiaoquan, a professor at Qingdao University and one senior author of the study, noting that the roadmap traces high similarities between microbiomes. “Although the compositions are distinct by habitat, each microbiome is, on average, only six “relatives” (or seven “steps”) from any other microbiome on Earth, indicating the inherent homology and common origin of the microbiomes at the global scale.”
On the other hand, information from the global MTN can indicate important information, such as evolvement or interaction of microbiomes. For example, the roadmap revealed that, the oceans are the most likely microbiomes that interact with beach sands and marine fishes, while soil and fresh water are the gateway of microbial exchange between the environment and plants or humans.
With the rapid change in climate as well as civilization on Earth, numerous microbiomes are disappearing, and emerging, every day. Although only a tiny proportion of them were recorded via metagenome sequencing, to construct and update the global MTN can be a “mission impossible”, without powerful search engines and standardized databases for microbiomes such as those in MSE.
“Such search-based global microbiome networks, reconstructed within hours on just one computing mode via MSE, provide a readily expanded reference for tracing the origin and evolution of existing microbiomes, and perhaps for guiding the design of new microbiomes” said XU Jian, Director of Single-Cell Center at QIBEBT and the other senior author of the study.
###
The CAS Microbiome Initiative, the National Natural Science Foundation of China, U.S. National Institutes of Health, U.S. National Science Foundation and the Alfred P. Sloan Foundation supported this work.
Media Contact
CHENG Jing
[email protected]
Related Journal Article
http://dx.