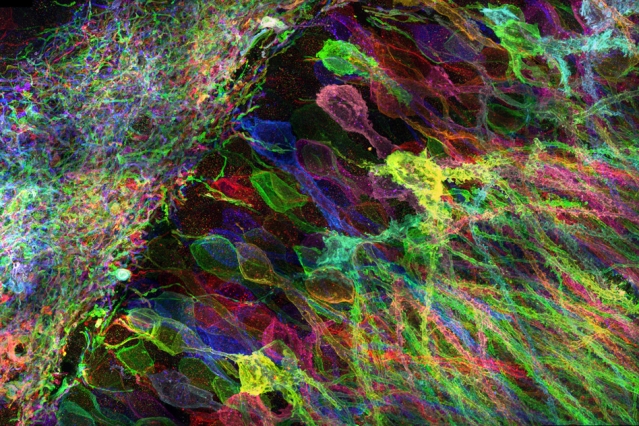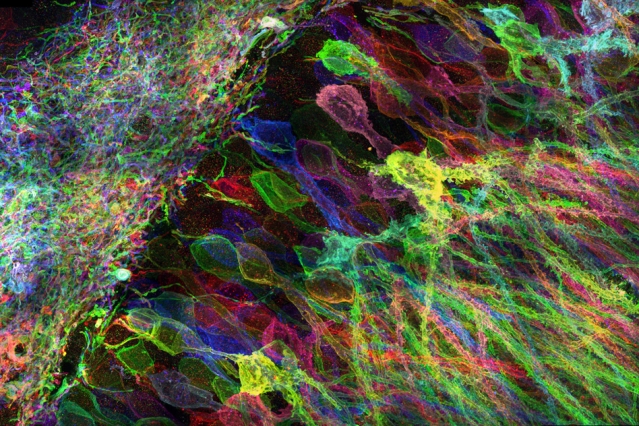
Cells contain thousands of messenger RNA molecules, which carry copies of DNA’s genetic instructions to the rest of the cell. MIT engineers have now developed a way to visualize these molecules in higher resolution than previously possible in intact tissues, allowing researchers to precisely map the location of RNA throughout cells.
Key to the new technique is expanding the tissue before imaging it. By making the sample physically larger, it can be imaged with very high resolution using ordinary microscopes commonly found in research labs.
“Now we can image RNA with great spatial precision, thanks to the expansion process, and we also can do it more easily in large intact tissues,” says Ed Boyden, an associate professor of biological engineering and brain and cognitive sciences at MIT, a member of MIT’s Media Lab and McGovern Institute for Brain Research, and the senior author of a paper describing the technique in the July 4 issue of Nature Methods.
Microscopy imaging of hippocampal brain circuitry.
Video: Yosuke Bando, Fei Chen, Dawen Cai, Ed Boyden, and Young Gyu
Studying the distribution of RNA inside cells could help scientists learn more about how cells control their gene expression and could also allow them to investigate diseases thought to be caused by failure of RNA to move to the correct location.
Boyden and colleagues first described the underlying technique, known as expansion microscopy (ExM), last year, when they used it to image proteins inside large samples of brain tissue. In a paper appearing in Nature Biotechnology on July 4, the MIT team has now presented a new version of the technology that employs off-the-shelf chemicals, making it easier for researchers to use.
MIT graduate students Fei Chen and Asmamaw Wassie are the lead authors of the Nature Methods paper, and Chen and graduate student Paul Tillberg are the lead authors of the Nature Biotechnology paper.
A simpler process
The original expansion microscopy technique is based on embedding tissue samples in a polymer that swells when water is added. This tissue enlargement allows researchers to obtain images with a resolution of around 70 nanometers, which was previously possible only with very specialized and expensive microscopes.
However, that method posed some challenges because it requires generating a complicated chemical tag consisting of an antibody that targets a specific protein, linked to both a fluorescent dye and a chemical anchor that attaches the whole complex to a highly absorbent polymer known as polyacrylate. Once the targets are labeled, the researchers break down the proteins that hold the tissue sample together, allowing it to expand uniformly as the polyacrylate gel swells.
In their new studies, to eliminate the need for custom-designed labels, the researchers used a different molecule to anchor the targets to the gel before digestion. This molecule, which the researchers dubbed AcX, is commercially available and therefore makes the process much simpler.
AcX can be modified to anchor either proteins or RNA to the gel. In the Nature Biotechnology study, the researchers used it to anchor proteins, and they also showed that the technique works on tissue that has been previously labeled with either fluorescent antibodies or proteins such as green fluorescent protein (GFP).
“This lets you use completely off-the-shelf parts, which means that it can integrate very easily into existing workflows,” Tillberg says. “We think that it’s going to lower the barrier significantly for people to use the technique compared to the original ExM.”
Using this approach, it takes about an hour to scan a piece of tissue 500 by 500 by 200 microns, using a light sheet fluorescence microscope. The researchers showed that this technique works for many types of tissues, including brain, pancreas, lung, and spleen.
Imaging RNA
In the Nature Methods paper, the researchers used the same kind of anchoring molecule but modified it to target RNA instead. All of the RNAs in the sample are anchored to the gel, so they stay in their original locations throughout the digestion and expansion process.
After the tissue is expanded, the researchers label specific RNA molecules using a process known as fluorescence in situ hybridization (FISH), which was originally developed in the early 1980s and is widely used. This allows researchers to visualize the location of specific RNA molecules at high resolution, in three dimensions, in large tissue samples.
This enhanced spatial precision could allow scientists to explore many questions about how RNA contributes to cellular function. For example, a longstanding question in neuroscience is how neurons rapidly change the strength of their connections to store new memories or skills. One hypothesis is that RNA molecules encoding proteins necessary for plasticity are stored in cell compartments close to the synapses, poised to be translated into proteins when needed.
With the new system, it should be possible to determine exactly which RNA molecules are located near the synapses, waiting to be translated.
“People have found hundreds of these locally translated RNAs, but it’s hard to know where exactly they are and what they’re doing,” Chen says. “This technique would be useful to study that.”
Boyden’s lab is also interested in using this technology to trace the connections between neurons and to classify different subtypes of neurons based on which genes they are expressing.
The research was funded by the Open Philanthropy Project, the New York Stem Cell Foundation Robertson Award, the National Institutes of Health, the National Science Foundation, and Jeremy and Joyce Wertheimer.
Story Source:
The above post is reprinted from materials provided by MIT NEWS