In a groundbreaking study that intertwines serendipity with rigorous scientific investigation, researchers at Georgia Tech have shed new light on the evolutionary processes driven by whole-genome duplication (WGD). This complex phenomenon, which involves the replication of an organism’s entire DNA, frequently propels significant evolutionary adaptations, although its mechanics have historically remained elusive. The study led by William Ratcliff and his collaborator, Kai Tong, explores not just the occurrence but also the stability of WGD over prolonged evolutionary periods, offering fresh insights into how genetic duplication underpins the emergence of multicellularity.
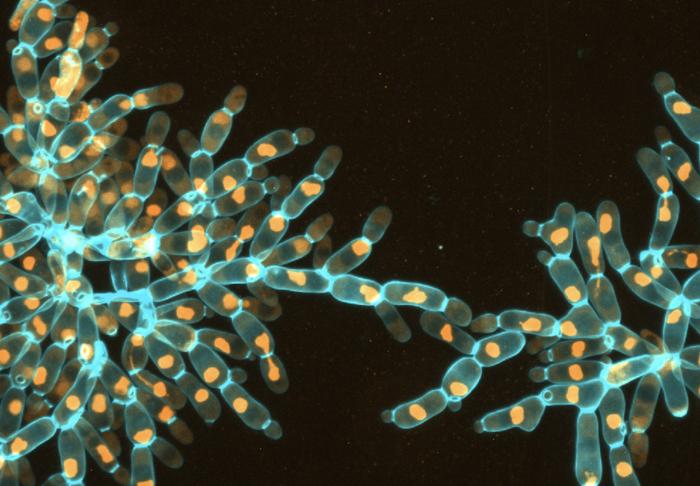
The research emerged from a broader inquiry into the transition of singular cells to multicellular organisms, a subject that has captivated biological scientists for decades. The team implemented the Multicellular Long-Term Evolution Experiment (MuLTEE), targeting the evolution of “snowflake” yeast, specifically Saccharomyces cerevisiae. These yeast strains are inherently ideal for studying evolutionary dynamics due to their rapid growth and ability to exhibit multicellularity as a response to selective pressures. This controlled environment allowed the scientists to observe and evaluate how WGD can arise and flourish under sustained, rigorous conditions.
Previous studies had suggested that WGD tends to be an unstable state, with organisms reverting back to their original diploid forms—those carrying two sets of chromosomes—within a few generations. This instability posed significant questions about the evolutionary value of tetraploids, which possess four chromosome sets. However, in a remarkable twist, it was found that the yeast retained their tetraploid status for over a thousand generations. This extended persistence marks a world-first observation of spontaneous WGD maintaining stability in laboratory conditions, thus challenging established tenets of evolutionary biology.
At the heart of their findings lies a surprising adaptability. The research revealed that by duplicating their genomes early in the experiment, within just 50 days, these snowflake yeast cells gained a critical advantage: larger cell sizes facilitated the formation of robust multicellular clusters. These clusters, in turn, thrived under the size selection imposed by the MuLTEE methodology. By continuing to select for larger cellular structures, the team inadvertently created a favorable environment for tetraploidy to not only arise but to be actively selected for over extended evolutionary timelines.
The mechanism that allows for this extended stability was revealed through exhaustive genetic analyses, which connected cellular and genomic changes to the enhanced fitness of larger multicellular assemblies. As the yeast underwent aneuploidy—a condition relevant to the abnormal number of chromosomes—the resulting genetic variation contributed significantly to the growth of complex multicellularity and survival advantages previously unobserved in microbial life.
Moreover, as the study unfolded, the collaborative efforts of undergraduate researchers became a testament to the value of hands-on exploration in scientific education. The active participation of students defined the experimental progress, affirming that authentic research experiences are indeed pivotal for budding scientists. The work of students like Vivian Cheng, who genetically engineered various yeast strains, directly contributed to the analytical findings central to understanding WGD’s role in evolutionary innovation.
This unexpected discovery highlights a fundamental aspect of scientific research: many significant breakthroughs stem from paths previously uncharted. The team’s observations underline that, as scientists delve into the unknown, unanticipated findings can yield insights that not only expand the horizon of current understanding but radically reshape our view of nature’s evolutionary tapestry.
The implications of this study stretch far beyond the immediate research context. The persistence of WGD in laboratory conditions opens up broader questions about the role of genetic duplication in evolutionary history across diverse organisms. It suggests that similar mechanisms may have played critical roles during pivotal points in evolutionary time, potentially explaining episodes of sudden biological complexity seen in the fossil record.
As the scientific community absorbs these findings, it is poised for future explorations that may further elucidate the nuances of evolution—especially regarding how organisms respond to selection pressures through genomic flexibility. This exploration constitutes a modern interpretation of Darwinian principles, reiterating that biological evolution is less about a linear progression and more about an intricate dance of genetics, environment, and chance.
In summary, the groundbreaking research from Georgia Tech offers vital new perspectives on evolutionary mechanisms, demonstrating that unexpected discoveries can lead to profound advancements in understanding biological complexity. As further studies build upon this foundational work, scientists may uncover even more about the interconnected pathways of life, revealing the unfolding narrative of evolution with each new twist.
While the road to scientific discovery is often complex and winding, it is in these unexpected intersections that the most substantial revelations emerge, driving the field forward. With continued curiosity and determination, researchers will certainly unlock more secrets that life and evolution hold, paving the way for innovative tests and theories in the biological sciences.
Subject of Research: Whole-genome duplication and its role in evolutionary biology
Article Title: Genome duplication in a long-term multicellularity evolution experiment
News Publication Date: 5-Mar-2025
Web References: Nature Journal
References: Tong, K., Datta, S., Cheng, V. et al. Genome duplication in a long-term multicellularity evolution experiment. Nature (2025).
Image Credits: Credit: Georgia Institute of Technology
Keywords: Evolutionary biology, genome duplication, snowflake yeast, tetraploidy, aneuploidy, multicellularity, experimental evolution, genetic research, biological complexity, undergraduate research, science education.
Tags: evolutionary adaptation mechanismsevolutionary dynamics in yeastgenetic duplication and stabilityimpact of WGD on adaptationlong-term evolutionary processesMulticellular Long-Term Evolution Experimentmulticellular organism evolutionrigorous scientific investigationserendipity in scientific discoverysnowflake yeast Saccharomyces cerevisiaetransition from unicellularity to multicellularitywhole-genome duplication