In the realm of cellular biology, understanding the intricate dynamics of how cells proliferate, differentiate, and respond to environmental stimuli has long been a scientific priority. Despite advanced molecular techniques, the mechanisms that underlie cellular heterogeneity—how genetically identical cells can exhibit diverse behaviors and fates—remain largely elusive. Recent pioneering research from the University of Zurich (UZH) breaks new ground by offering an unprecedented view into the multigenerational development of cells, particularly cancer cells, using cutting-edge live-cell tracking and genome editing technologies. This work not only unfolds the biological complexity hidden in cell lineages but also reveals critical implications for cancer progression and therapeutic resistance.
Cells, as the fundamental units of life, encompass an extraordinary diversity even within ostensibly uniform populations such as tissues or tumor masses. This diversity, or heterogeneity, stems from both genetic mutations that alter DNA sequences and epigenetic modifications that influence gene expression without changing the underlying code. These layers of complexity generate a mosaic of cell behaviors that enable development, adaptation to stress, and, conversely, contribute to disease states such as cancer. The new study led by UZH researchers leverages advanced CRISPR-based genome editing to probe these phenomena with remarkable temporal and spatial resolution.
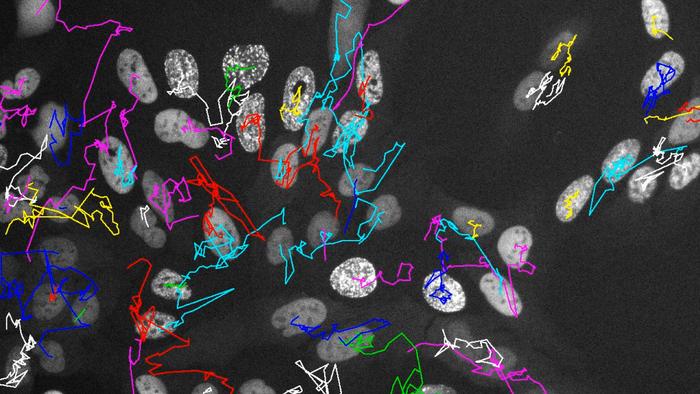
Central to this breakthrough is the integration of fluorescent markers fused to key proteins involved in DNA replication and the DNA damage response. By genetically engineering cells to express these markers, researchers could visualize and quantitatively track the dynamics of genome duplication and accumulation of heritable DNA damage in living cells through successive generations. This approach surpasses traditional snapshot assays by permitting real-time follow-up of the same cellular lineages as they evolve, divide, and differentiate under various stress conditions.
Through sophisticated microscopy combined with computer-assisted tracking software, the team followed the progeny of individual cancer cells, capturing the onset, progression, and resolution of DNA replication events alongside DNA damage signals. One of the most striking findings was that after exposure to stress, daughter cells derived from a single mother cell no longer behaved synchronously. Instead, they exhibited pronounced divergences in DNA replication timing and cell cycle regulation, indicating that stress-induced perturbations are propagated—and even amplified—across multiple generations.
This desynchronization among sibling cells under stress sheds light on the mechanisms by which cellular heterogeneity is established and maintained within tumors. Differences in DNA replication kinetics and cell cycle protein production suggest that cells may adopt divergent trajectories, potentially leading to subpopulations with distinct functional capacities and differential responses to therapies. Such intratumoral heterogeneity is a recognized obstacle to effective cancer treatment, as resistant clones can emerge and drive relapse.
Delving deeper, the study elucidates the origins of polyploidy—a state wherein cells acquire multiple copies of the entire genome. Polyploidy notably increases genomic complexity and is frequently observed in aggressive cancers, where it facilitates adaptation and survival. With the aid of multigenerational tracking, the UZH group identified several pathways leading to polyploidy, each imparting unique effects on genomic stability and cell fitness. These insights underscore that not all polyploidization events are equivalent; their specific mechanisms could influence the evolutionary trajectories of cancer cell populations.
Importantly, the comprehensive integration of real-time imaging data with endpoint molecular analyses allowed the researchers to correlate dynamic processes with lasting cellular outcomes. DNA damage incurred in one generation was found to have heritable effects manifested in later progeny, reinforcing the concept that stress responses are not transient but shape the phenotypic landscape over time. These findings challenge the traditional view of cell lineages as mere replicates and position them as dynamic entities with cumulative memory of past insults.
From a methodological standpoint, the fusion of CRISPR-mediated genome editing with advanced live-cell imaging represents a powerful platform for future studies. It enables high-resolution dissection of complex biological phenomena beyond bulk measurements, capturing single-cell nuances essential for understanding development, disease progression, and therapeutic resistance. The researchers also highlight the potential for automation and artificial intelligence to manage and interpret the vast data generated by such high-throughput single-cell analyses, an indispensable advance given the scale of cellular heterogeneity.
The implications of this work extend beyond academic interest. By defining how genetic and epigenetic heterogeneity develop and persist, the study paves the way for interventions that could manipulate these processes to improve cancer treatment. For example, controlling the pathways leading to polyploidy or enhancing the fidelity of DNA damage responses might render tumor cells more vulnerable to existing therapies. Tailoring treatments based on lineage-specific vulnerabilities could represent a paradigm shift toward more personalized and effective oncology.
This research marks a significant milestone, revealing just the cusp of a deeper understanding of cellular complexity. As Professor Matthias Altmeyer and his team continue refining their methodologies, the promise of unraveling the “tip of the iceberg” becomes ever more tangible. The marriage of molecular genetics, live-cell imaging, and computational analysis is set to transform our grasp of biology at its most fundamental level, with profound repercussions for medicine and biotechnology.
In conclusion, the UZH study offers a detailed blueprint of how cellular heterogeneity arises and is perpetuated through generations, spotlighting the intricate interplay between DNA replication fidelity, damage inheritance, and stress responses. It challenges existing paradigms by demonstrating heritability of stress-induced variations and elucidates the multifaceted origins of polyploidy linked to cancer resilience. This innovative approach sets a new standard for cellular lineage tracing and provides an invaluable framework for future research aiming to conquer the complexities of tumor evolution and therapeutic resistance.
—
Subject of Research: Cells
Article Title: Andreas Panagopoulos, Merula Stout et al. Multigenerational cell tracking of DNA replication and heritable DNA damage. Nature. 21 May 2025. DOI: 10.1038/s41586-025-08986-0
News Publication Date: 21-May-2025
Web References: http://dx.doi.org/10.1038/s41586-025-08986-0
References: Andreas Panagopoulos, Merula Stout et al. Multigenerational cell tracking of DNA replication and heritable DNA damage. Nature. 21 May 2025. DOI: 10.1038/s41586-025-08986-0
Image Credits: Andreas Panagopoulos, Merula Stout et al.
Keywords: Tumor cells, Cancer cell lines, Live cells, Daughter cells, Molecular genetics, DNA damage, DNA replication, Genetic engineering
Tags: adaptation of cells to environmental stimulicellular heterogeneity in cancerepigenetic modifications and gene expressionfundamental units of life in biologygenetic mutations in cellular diversitygenome editing in cancer researchlive-cell tracking technologiesmultigenerational cell developmentreal-time insights into cancer cellsstress impact on cell behaviortherapeutic resistance in cancer therapiesUZH cancer research advancements