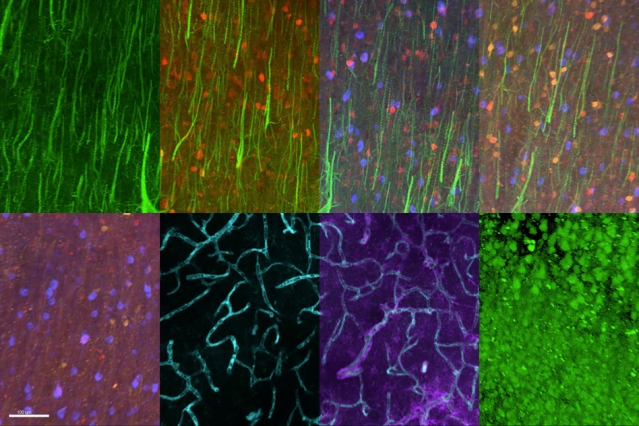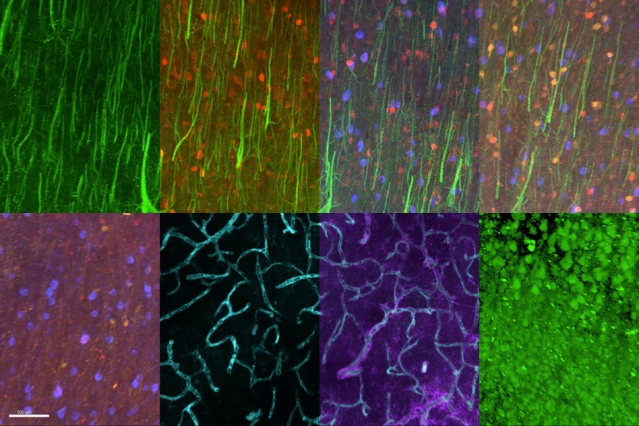
MIT chemical engineers and neuroscientists have developed a new way to classify neurons by labeling and imaging the proteins found in each cell. This type of imaging offers clues to each neuron’s function and should help in mapping the human brain, the researchers say.
“Each cell uses a unique combination of proteins. It’s basically a fingerprint,” says Kwanghun Chung, who is the Samuel A. Goldblith Assistant Professor in the Department of Chemical Engineering, a member of MIT’s Institute for Medical Engineering and Science (IMES) and Picower Institute for Learning and Memory, and the leader of the research team. “If you can look at expression patterns of many proteins, then you can guess each cell’s type and what it’s doing.”
Using this approach, the researchers were able to visualize 22 different proteins inside human brain slices, but the method could be scaled to analyze many more proteins and larger tissue samples. This could help scientists learn more about how diseases alter brain chemistry.
“Now, researchers will be able to investigate the differences between brains from disease models and normal animals, simultaneously looking at potentially dozens of different molecules. This is very important, as the individual variation between brains would make it difficult to make solid connections when looking at those same molecules, one at a time, between dozens of samples,” says graduate student Evan Murray, one of the lead authors of a paper describing the technique in the Dec. 3 issue of Cell.
The paper’s other lead authors are graduate students Jae Hun Cho, Daniel Goodwin, and Justin Swaney, and postdoc Taeyun Ku.
Using a novel method, researchers are able to image and label proteins found in each brain cell from a single tissue sample.
Video: Melanie Gonick/MIT (protein imaging renderings courtesy of Kwanghun Chung and Evan Murray)
Label, rinse, repeat
The key advance of the new technology, known as SWITCH, is the ability to preserve tissue in such a way that it can be imaged repeatedly, with different proteins labeled each time.
To achieve that, the researchers devised a method for controlling the chemical reactions required for tissue preservation and labeling. This allows them to first preserve the tissue, then label a certain protein and image it. They can then wash away the tagging molecule and label a different protein, over and over again.
Controlling the chemical reactions requires a pair of buffers — solutions of weak acids and bases — that alter the tissue’s environment. One of the buffers, known as SWITCH-Off, halts most chemical reactions in the tissue, while the SWITCH-On buffer allows them to resume.
To prepare the tissue samples, the researchers first add the SWITCH-Off buffer, followed by chemicals necessary for tissue preservation, the most important of which is glutaraldehyde. Because the chemicals cannot react with any cells, they diffuse evenly throughout the sample. “It’s like these chemicals are in a stealth mode. They are not detected by tissue,” Chung says.
When the researchers add the SWITCH-On buffer, the glutaraldehyde forms a gel that preserves the tissue. The researchers also add detergent to destroy the lipids of the cell membranes, making the cell interiors more visible to a light microscope.
Once the tissue is preserved and ready for imaging, the researchers add the SWITCH-Off buffer again. With the tissue in an unreactive state, they add labels such as antibodies or dyes, which can be tailored to detect not only proteins but also DNA, neurotransmitters, or lipids. Once the labels have diffused through the tissue, adding the SWITCH-On buffer allows all cells to be exposed to the labels simultaneously.
Protein analysis
In the Cell study, the researchers labeled 22 different proteins in a small section of human brain tissue (roughly 3 millimeters by 3 millimeters by 0.1 millimeters). After 22 rounds of labeling, the tissue was still in good condition, so the researchers believe this technique could be used to image even more proteins.
They also examined the distribution of six proteins in human visual cortex tissue and were able to label and image the myelinated fibers that connect different regions of the brain. “If you can visualize these fibers then you can really understand brain connectivity and the fundamental laws that govern how these wires are formed and connected,” Chung says.
The size of the tissue that can be imaged is limited only by the amount of time required for labeling the proteins and imaging the sample.
It takes about a month for each labeling molecule to diffuse through a cubic-centimeter-sized tissue sample, but Chung and colleagues recently reported in the Proceedings of the National Academy of Sciences that they could speed this up dramatically by exposing the tissue to a randomly changing electric field. This cuts the diffusion time to about a day.
The imaging time depends on the type of microscope used. For this study, the researchers used a light sheet microscope, which can image samples about 100 times faster than a traditional light microscope. Using this microscope, it took about two hours to image an entire mouse brain, compared to about three days with a traditional microscope.
“There are other ways of doing proteomic imaging, but many of them are two-dimensional, or not scalable, or require special equipment,” Chung says. “But with this technique, anyone can do it and it’s scalable.”
Robert Brown, chair of neurology at the University of Massachusetts Medical School, says the new technique is part of a “new generation of imaging technology based on careful manipulation of biochemical structures.”
“It’s extraordinary because it allows one to look for multiple targets simultaneously in the same cell, with three-dimensional resolution, which has not been feasible with previous imaging methods,” he adds.
Chung’s lab will make detailed protocols and other resources available through its website. He now plans to start using SWITCH to study human neurological disorders and is also working on other technologies to help map the human brain.
Story Source:
The above post is reprinted from materials provided by MIT NEWS
The post Protein imaging reveals detailed brain architecture appeared first on Scienmag.