Credit: OIST
Unlike experimental neuroscientists who deal with real-life neurons, computational neuroscientists use model simulations to investigate how the brain functions. While many computational neuroscientists use simplified mathematical models of neurons, researchers in the Computational Neuroscience Unit at the Okinawa Institute of Science and Technology Graduate University (OIST) develop software that models neurons to the detail of molecular interactions with the goal of eliciting new insights into neuronal function. Applications of the software were limited in scope up until now because of the intense computational power required for such detailed neuronal models, but recently Dr. Weiliang Chen, Dr. Iain Hepburn, and Professor Erik De Schutter published two related papers in which they outline the accuracy and scalability of their new high-speed computational software, "Parallel STEPS". The combined findings suggest that Parallel STEPS could be used to reveal new insights into how individual neurons function and communicate with each other.
The first paper, published in The Journal of Chemical Physics in August 2016, focusses on ensuring that the accuracy of Parallel STEPS is comparable with conventional methods. In conventional approaches, computations associate with neuronal chemical reactions and molecule diffusion are all calculated on one computational processing unit or 'core' sequentially. However, Dr. Iain Hepburn and colleagues introduced a new approach to perform computations of reaction and diffusion in parallel which can then be distributed over multiple computer cores, whilst maintaining simulation accuracy to a high degree. The key was to develop an original algorithm separated into two parts – one that computed chemical reaction events and the other diffusion events.
"We tested a range of model simulations from simple diffusion models to realistic biological models and found that we could achieve improved performance using a parallel approach with minimal loss of accuracy. This demonstrated the potential suitability of the method on a larger scale," says Dr. Hepburn.
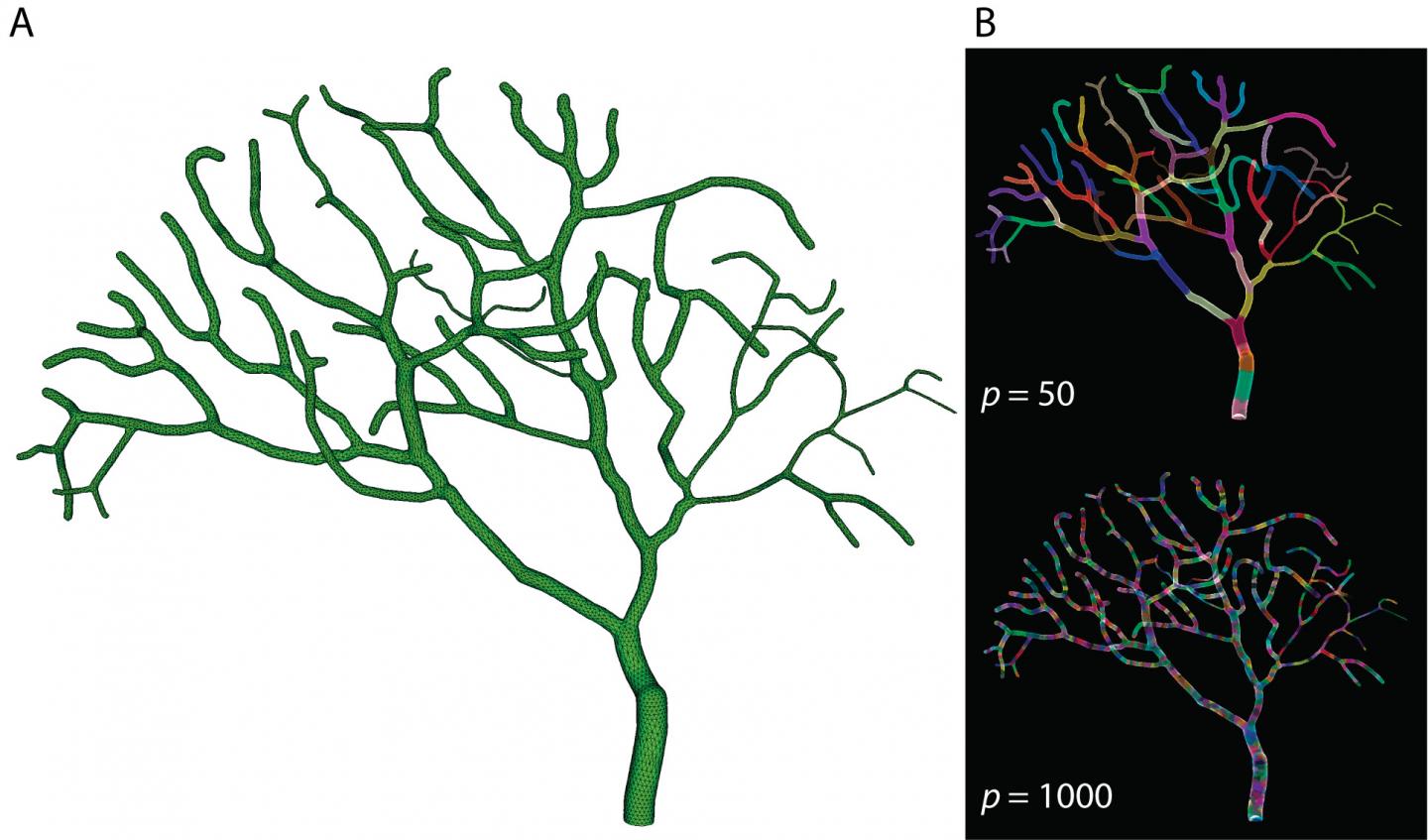
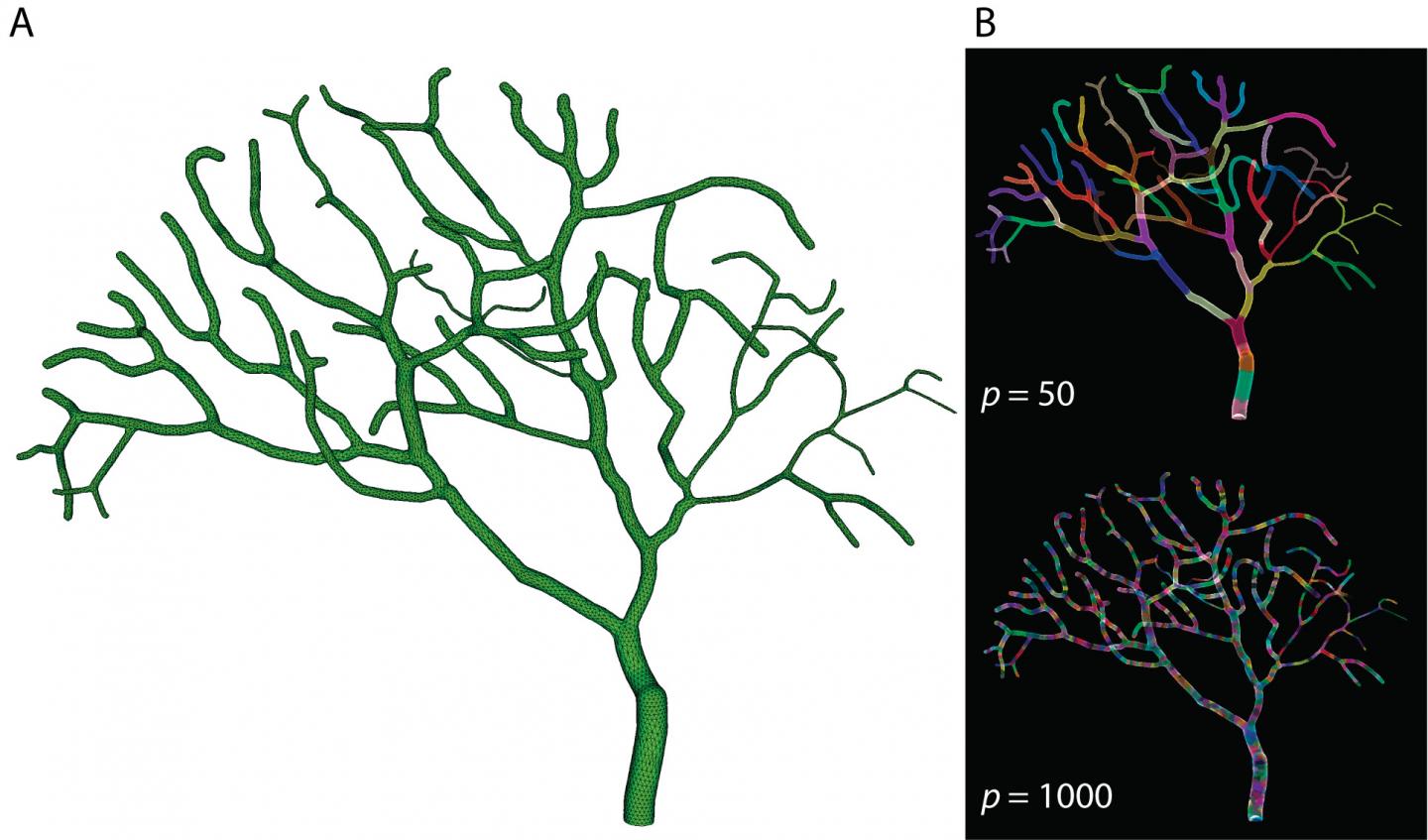
In a related paper published in Frontiers in Neuroinformatics this February, Dr. Weiliang Chen presented the implementation details of Parallel STEPS and investigated its performance and potential applications. By breaking a partial model of a Purkinje cell – one of the largest neurons in the brain – into 50 to 1000 sections and simulating reaction and diffusion events for each section in parallel on the Sango supercomputer at OIST, Dr. Chen and colleagues saw dramatically increased computation speeds. They tested this approach on both simple models and more complicated models of calcium bursts in Purkinje cells and demonstrated that parallel simulation could speed up computations by more than several hundred times that of conventional methods.
"Together, our findings show that Parallel STEPS implementation achieves significant improvements in performance, and good scalability," says Dr. Chen. "Similar models that previously required months of simulation can now be completed within hours or minutes, meaning that we can develop and simulate more complex models, and learn more about the brain in a shorter amount of time."
Dr. Hepburn and Dr. Chen from OIST's Computational Neuroscience Unit, led by Professor Erik De Schutter, are actively collaborating with the Human Brain Project, a world-wide initiative based at École Polytechnique Fédérale de Lausanne (EPFL) in Switzerland, to develop a more robust version of Parallel STEPS that incorporates electric field simulation of cell membranes.
So far STEPS is only realistically capable of modeling parts of neurons but with the support of Parallel STEPS, the Computational Neuroscience Unit hopes to develop a full-scale model of a whole neuron and subsequently the interactions between neurons in a network. By collaborating with the EPFL team and by making use of the IBM 'Blue Gene/Q' supercomputer located there, they aim to achieve these goals in the near future.
"Thanks to modern supercomputers we can study molecular events within neurons in a much more transparent way than before," says Prof. De Schutter. "Our research opens up interesting avenues in computational neuroscience that links biochemistry with electrophysiology for the first time."
###
Media Contact
Kaoru Natori
[email protected]
899-896-62389
@oistedu
http://www.oist.jp/
############
Story Source: Materials provided by Scienmag