A joint Russian-French-German paper was published in Nature Communications
Credit: Kazan Federal University
The Russian side is represented by Structural Biology Lab (Kazan Federal University) and Institute of Proteins (Russian Academy of Sciences). This particular paper tackles the issue of stress resistance in Staphylococcus aureus. The results can help in finding new antibiotics.
Head of Structural Biology Lab Konstantin Usachev explains that this was the fruit of a five-year-long cooperation between KFU, Institute of Proteins, Stuttgart University, and Institute of Genetics and Molecular and Cellular Biology (Strasbourg, France). In 2016, the team was first to completely describe the structure of Staphylococcus aureus ribosome and compare it to other organisms.
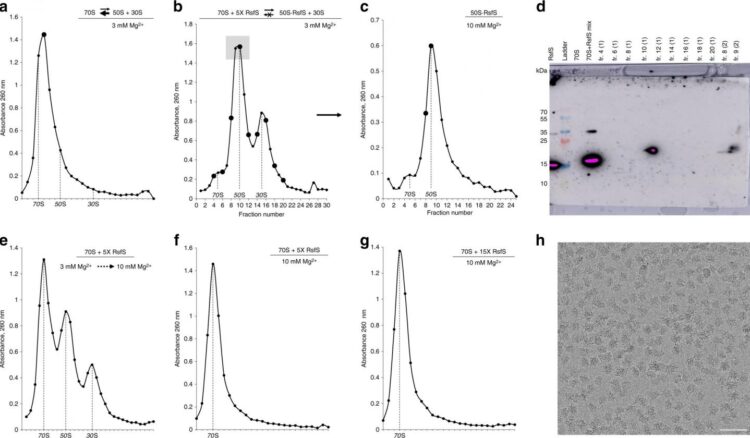
“The ribosome is the largest ribonucleic complex in the cell, and it consists of two subunits: large and small. The small subunit is responsible for reading the genetic code, and the function of the large subunit is to ensure the formation of the peptide bond in the growing protein chain. In our article, using cryoelectron microscopy and X-ray diffraction methods, we were able to show the ribosome binding mechanism of the RsfS protein (Ribosome silencing factor S), which protects Staphylococcus aureus from stress (antibiotics, fever, or host immunity). Under stress, this protein binds to the large subunit of the ribosome and prevents the small subunit from joining, preventing the formation of functional ribosomes,” says Usachev.
Research in this area began 5 years ago. For a long time, scientists were not able to obtain the structure of a complex of bacterial ribosomes with the RsfS protein in high resolution, which was necessary to understand the details of its mechanism of action.
“One of the problems was the high toxicity of this protein to the cells of the bacteria E. coli, the organism used to produce proteins in the laboratory. The fact is that the RsfS protein, which stops the synthesis of proteins in Staphylococcus aureus, is able to stop this process in other bacteria. This resulted in a very small amount of protein sample, insufficient for structural studies. In addition, the sample was extremely unstable and aggregated. Then the idea came to us – to isolate this protein simultaneously with its target in the structure of staphylococcus ribosome – protein L14, which is a part of the large subunit. It turned out that if you select both components at the same time, they will be stable in solution. We were able to obtain crystals of these proteins and solve the structure by X-ray diffraction analysis, first with medium resolution using the new single-crystal diffractometer available in our laboratory, and then with high resolution using the ESRF synchrotron in Grenoble, France,” continues the interviewee.
Next, the scientists needed to study the details of the interaction of the RsfS protein with Staphylococcus aureus ribosome. This could be achieved with cryoelectron microscopy.
“Unfortunately, we did not have a microscope capable of solving high-resolution structures using this method, but NovAliX became interested in our studies and offered their microscope to test the initial samples. The obtained samples of the complexes had turned out so good that the company then contacted one of the world’s leading manufacturers of FEI microscopes in the Netherlands and organized data collection on a Titan Krios microscope. As a result, combining the data of cryoelectron microscopy with the previously obtained data of X-ray diffraction analysis, we were able to show in detail the molecular mechanism of the action of the RsfS protein on Staphylococcus aureus ribosomes,” concludes Usachev.
Currently, Structural Biology Lab is partnering up with the Chemoinformatics Lab to predict the structure of potential antibiotics disrupting the functioning of RsfS protein and thus effectively eliminating Staphylococcus.
According to Usachev, only combined efforts of biologists, chemists and physicists can advance the research in protein synthesis and new medications.
###
Media Contact
Yury Nurmeev
[email protected]
Original Source
https:/
Related Journal Article
http://dx.